Chapter 11 Missing data
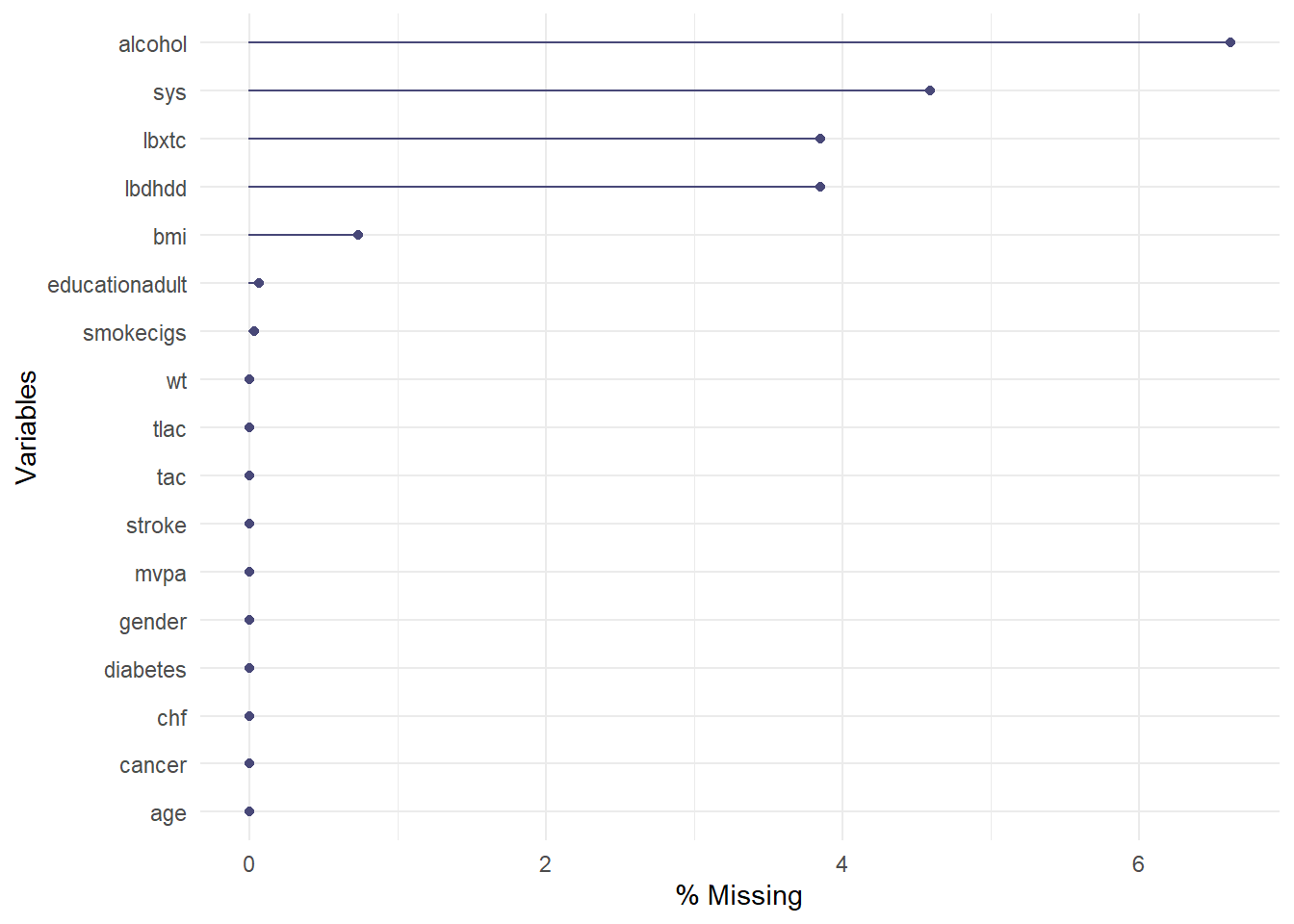
11.1 Per variable missingness
Number and percentage of missing.
| Variable | Missing (count) | Missing (%) |
|---|---|---|
| alcohol | 395 | 6.61 |
| sys | 274 | 4.59 |
| lbxtc | 230 | 3.85 |
| lbdhdd | 230 | 3.85 |
| bmi | 44 | 0.74 |
| educationadult | 4 | 0.07 |
| smokecigs | 2 | 0.03 |
| age | 0 | 0.00 |
| gender | 0 | 0.00 |
| diabetes | 0 | 0.00 |
| chf | 0 | 0.00 |
| cancer | 0 | 0.00 |
| stroke | 0 | 0.00 |
| tac | 0 | 0.00 |
| tlac | 0 | 0.00 |
| mvpa | 0 | 0.00 |
| wt | 0 | 0.00 |
Investigate for groups of variables:
| Variable | Missing (count) | Missing (%) |
|---|---|---|
| any_variable_missing | 768 | 12.86 |
| any_lab_missing | 470 | 7.87 |
| any_lifestyle_missing | 431 | 7.22 |
| any_demographics_missing | 4 | 0.07 |
| any_vip_missing | 0 | 0.00 |
| any_health_missing | 0 | 0.00 |
| any_physact_missing | 0 | 0.00 |
Overall, 13% are missing when all variables are included in a model. There is no missingness in the VIPs and 7% in life style variables such as smoking and alcohol consumption.
11.2 Variable summaries for complete vs incomplete cases
| complete (N=5204) | incomplete (N=768) | Total (N=5972) | p value | |
|---|---|---|---|---|
| age | 0.005 | |||
| Median | 54.167 | 50.417 | 53.750 | |
| Q1, Q3 | 41.917, 67.417 | 40.167, 66.167 | 41.646, 67.250 | |
| Range | 30.000 - 84.917 | 30.000 - 84.917 | 30.000 - 84.917 | |
| gender | < 0.001 | |||
| Male | 2601 (50.0%) | 334 (43.5%) | 2935 (49.1%) | |
| Female | 2603 (50.0%) | 434 (56.5%) | 3037 (50.9%) | |
| education level | < 0.001 | |||
| N-Miss | 0 | 4 | 4 | |
| Less than high school | 1431 (27.5%) | 252 (33.0%) | 1683 (28.2%) | |
| High school | 1251 (24.0%) | 197 (25.8%) | 1448 (24.3%) | |
| More than high school | 2522 (48.5%) | 315 (41.2%) | 2837 (47.5%) | |
| diabetes | 0.092 | |||
| No | 4558 (87.6%) | 656 (85.4%) | 5214 (87.3%) | |
| Yes | 646 (12.4%) | 112 (14.6%) | 758 (12.7%) | |
| congestive heart failure | 0.071 | |||
| No | 5010 (96.3%) | 729 (94.9%) | 5739 (96.1%) | |
| Yes | 194 (3.7%) | 39 (5.1%) | 233 (3.9%) | |
| cancer | 0.458 | |||
| No | 4664 (89.6%) | 695 (90.5%) | 5359 (89.7%) | |
| Yes | 540 (10.4%) | 73 (9.5%) | 613 (10.3%) | |
| stroke | 0.206 | |||
| No | 5003 (96.1%) | 731 (95.2%) | 5734 (96.0%) | |
| Yes | 201 (3.9%) | 37 (4.8%) | 238 (4.0%) | |
| body mass index | 0.003 | |||
| Median | 28.060 | 28.290 | 28.080 | |
| Q1, Q3 | 24.740, 32.180 | 24.432, 32.928 | 24.730, 32.230 | |
| Range | 13.360 - 130.210 | 14.650 - 63.420 | 13.360 - 130.210 | |
| smoking status | 0.006 | |||
| N-Miss | 0 | 2 | 2 | |
| Never | 2514 (48.3%) | 397 (51.8%) | 2911 (48.8%) | |
| Former | 1571 (30.2%) | 188 (24.5%) | 1759 (29.5%) | |
| Current | 1119 (21.5%) | 181 (23.6%) | 1300 (21.8%) | |
| alcohol consumption | 0.153 | |||
| Median | 1.000 | 2.000 | 1.000 | |
| Q1, Q3 | 1.000, 2.000 | 1.000, 2.000 | 1.000, 2.000 | |
| Range | 1.000 - 3.000 | 1.000 - 3.000 | 1.000 - 3.000 | |
| total log activity count (log(1+activity)) | < 0.001 | |||
| Median | 2925.685 | 2796.018 | 2910.926 | |
| Q1, Q3 | 2401.400, 3440.724 | 2239.750, 3380.564 | 2384.757, 3430.648 | |
| Range | 313.083 - 6122.678 | 466.036 - 5102.369 | 313.083 - 6122.678 | |
| total accelerometer wear time | 0.017 | |||
| Median | 854.310 | 832.100 | 852.071 | |
| Q1, Q3 | 785.000, 923.808 | 762.458, 909.571 | 782.851, 922.036 | |
| Range | 602.000 - 1440.000 | 600.000 - 1440.000 | 600.000 - 1440.000 |
11.3 Missingness patterns over variables
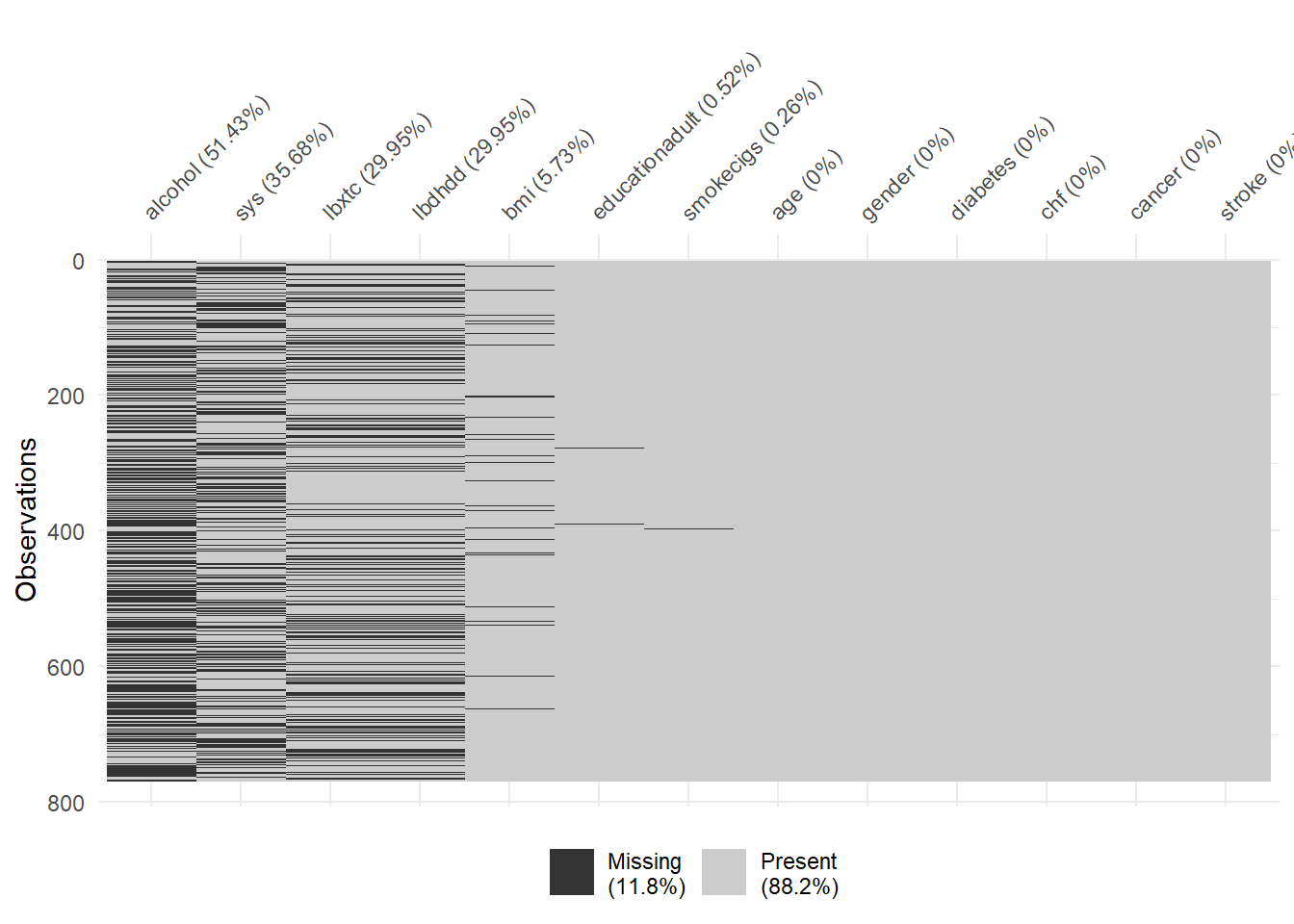
Missing values for each participant in the NHANES dataset is shown in the following figure, where the black lines correspond to observations with missing values.
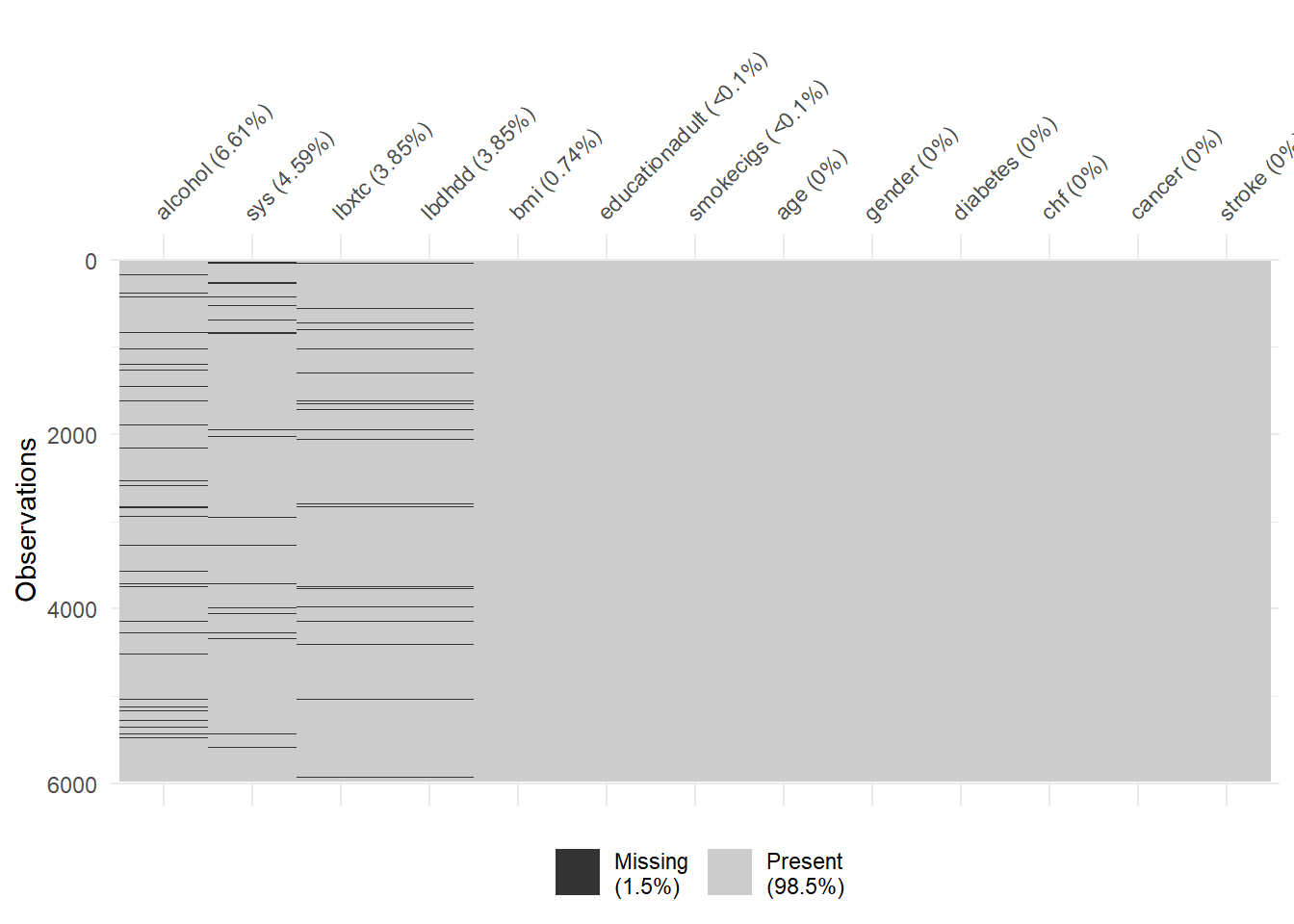
There are 7 independent variables with missing values in the dataset.
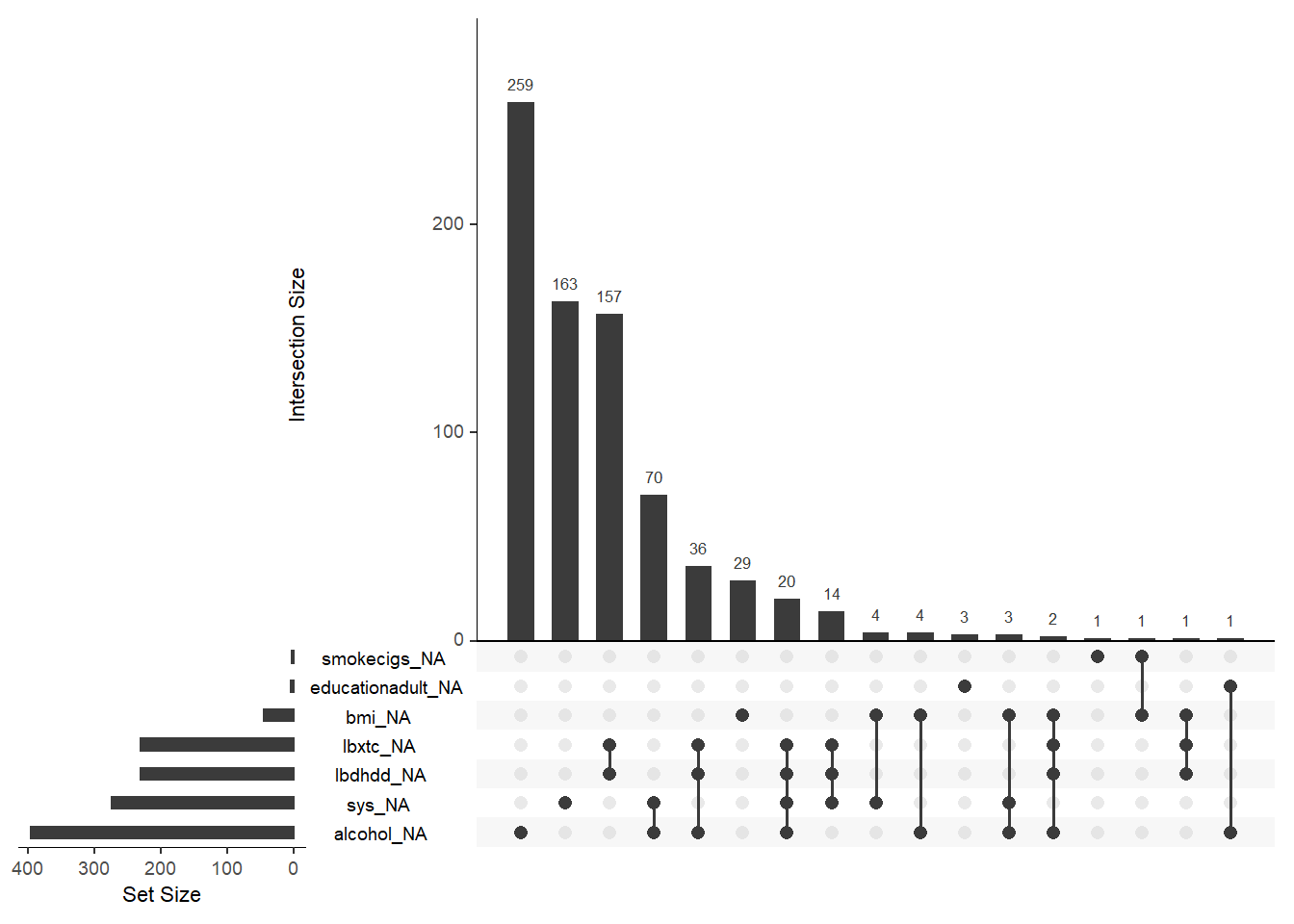
Physiological variables (blood pressure, cholesterol) have the highest proportion of missingness. There does not seem to be a pattern of missingness across variables, other than the cholesterol (toal, HDL) variables.
In addition, we can explore missing data mechanisms and relationships between BMI and systolic blood pressure, included in the same scatterplot:
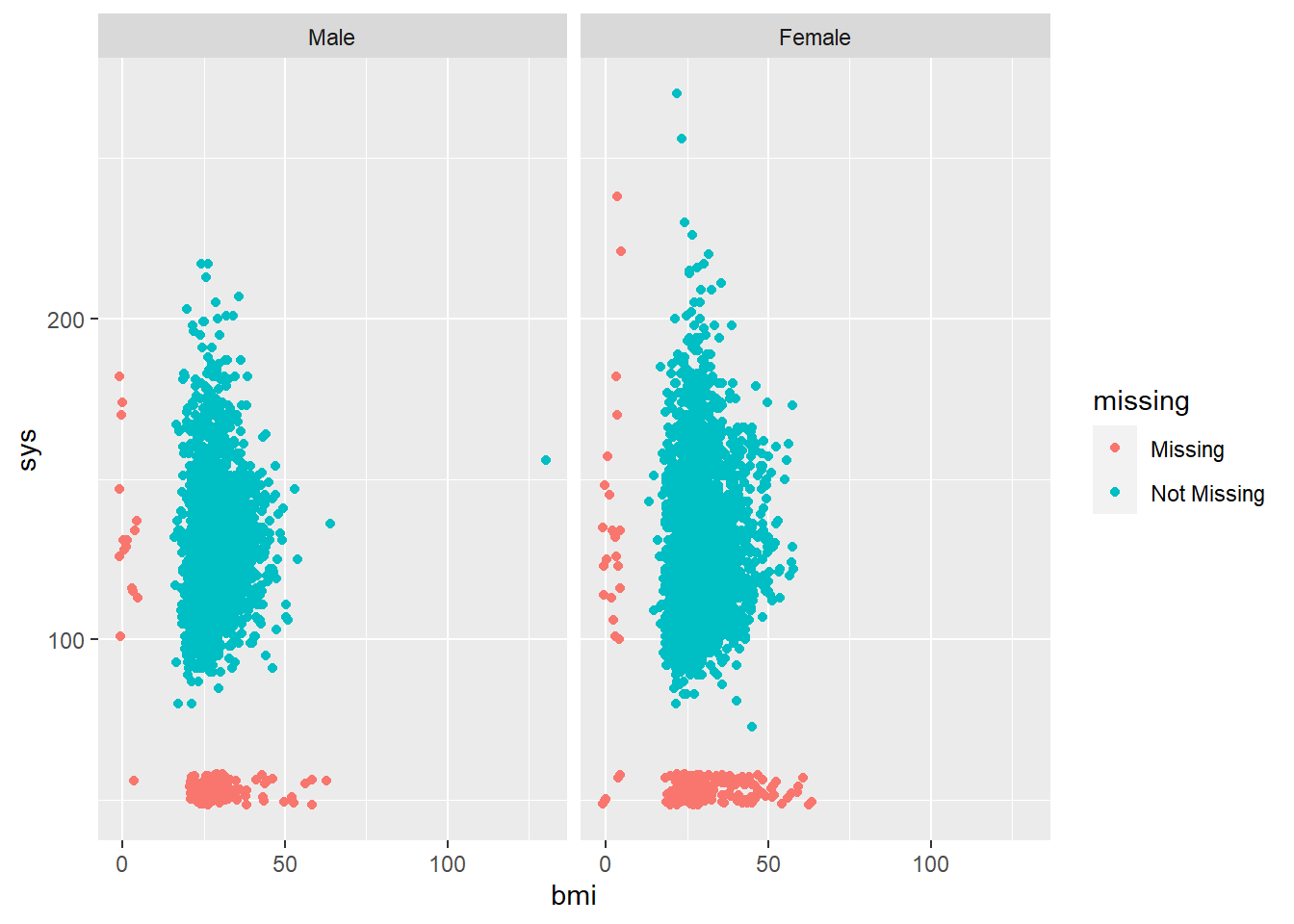
Missing values are seen across the same range of values for the other variable. There is an extreme value in BMI for males that is likely an entry error.
11.4 (In)complete cases
This section presents patients with a least one missing value. First we list out patients with at least one missing value in a filterable table.
Then we report the pattern of missing for this set of patients.
11.5 Section session info
## R version 4.1.3 (2022-03-10)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_Austria.1252 LC_CTYPE=English_Austria.1252
## [3] LC_MONETARY=English_Austria.1252 LC_NUMERIC=C
## [5] LC_TIME=English_Austria.1252
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] VIM_6.1.1 colorspace_2.0-3 arsenal_3.6.3 DT_0.22
## [5] kableExtra_1.3.4 gt_0.4.0 naniar_0.6.1 Hmisc_4.6-0
## [9] Formula_1.2-4 survival_3.2-13 lattice_0.20-45 forcats_0.5.1
## [13] stringr_1.4.0 dplyr_1.0.8 purrr_0.3.4 readr_2.1.2
## [17] tidyr_1.2.0 tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1
## [21] here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] ellipsis_0.3.2 class_7.3-20 visdat_0.5.3
## [4] rprojroot_2.0.2 htmlTable_2.4.0 base64enc_0.1-3
## [7] fs_1.5.2 rstudioapi_0.13 proxy_0.4-26
## [10] farver_2.1.0 fansi_1.0.3 lubridate_1.8.0
## [13] ranger_0.13.1 xml2_1.3.3 splines_4.1.3
## [16] robustbase_0.95-0 knitr_1.38 jsonlite_1.8.0
## [19] broom_0.7.12 cluster_2.1.2 dbplyr_2.1.1
## [22] png_0.1-7 compiler_4.1.3 httr_1.4.2
## [25] backports_1.4.1 assertthat_0.2.1 Matrix_1.4-0
## [28] fastmap_1.1.0 cli_3.2.0 htmltools_0.5.2
## [31] tools_4.1.3 gtable_0.3.0 glue_1.6.2
## [34] Rcpp_1.0.8.3 carData_3.0-5 cellranger_1.1.0
## [37] jquerylib_0.1.4 vctrs_0.3.8 svglite_2.1.0
## [40] crosstalk_1.2.0 lmtest_0.9-40 xfun_0.30
## [43] laeken_0.5.2 rvest_1.0.2 lifecycle_1.0.1
## [46] DEoptimR_1.0-11 zoo_1.8-9 MASS_7.3-55
## [49] scales_1.1.1 hms_1.1.1 RColorBrewer_1.1-2
## [52] yaml_2.3.5 gridExtra_2.3 UpSetR_1.4.0
## [55] sass_0.4.1 rpart_4.1.16 latticeExtra_0.6-29
## [58] stringi_1.7.6 highr_0.9 e1071_1.7-9
## [61] checkmate_2.0.0 boot_1.3-28 commonmark_1.8.0
## [64] rlang_1.0.2 pkgconfig_2.0.3 systemfonts_1.0.4
## [67] evaluate_0.15 labeling_0.4.2 htmlwidgets_1.5.4
## [70] tidyselect_1.1.2 plyr_1.8.7 magrittr_2.0.2
## [73] bookdown_0.25 R6_2.5.1 generics_0.1.2
## [76] DBI_1.1.2 pillar_1.7.0 haven_2.4.3
## [79] foreign_0.8-82 withr_2.5.0 abind_1.4-5
## [82] sp_1.4-6 nnet_7.3-17 modelr_0.1.8
## [85] crayon_1.5.1 car_3.0-12 utf8_1.2.2
## [88] tzdb_0.2.0 rmarkdown_2.13 jpeg_0.1-9
## [91] readxl_1.3.1 data.table_1.14.2 vcd_1.4-9
## [94] reprex_2.0.1 digest_0.6.29 webshot_0.5.2
## [97] munsell_0.5.0 viridisLite_0.4.0 bslib_0.3.1