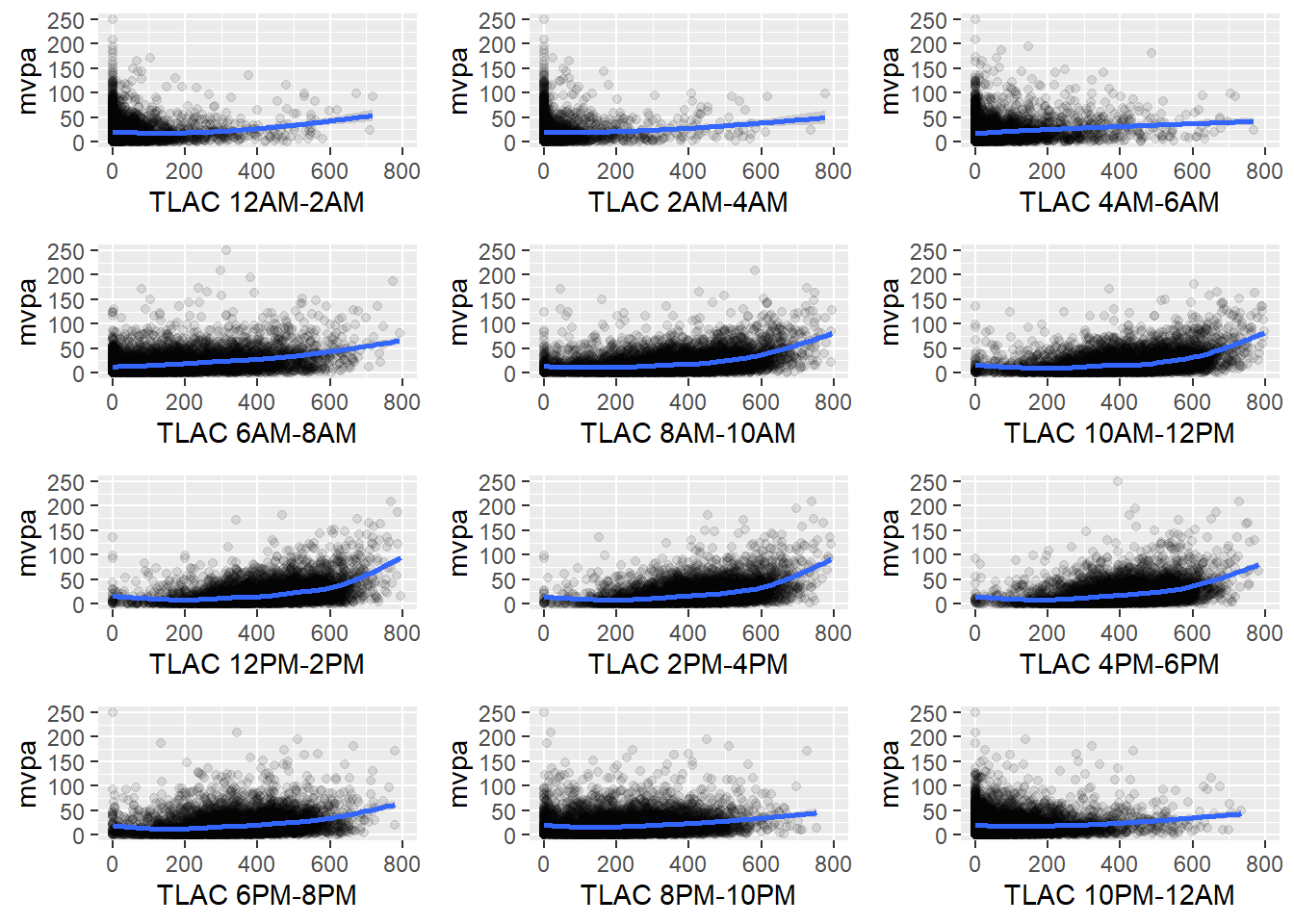
Chapter 13 Multivariate distributions
This section reports a series of multivariate summaries of the NHANES dataset.
13.1 Overview
13.1.1 Variable correlation
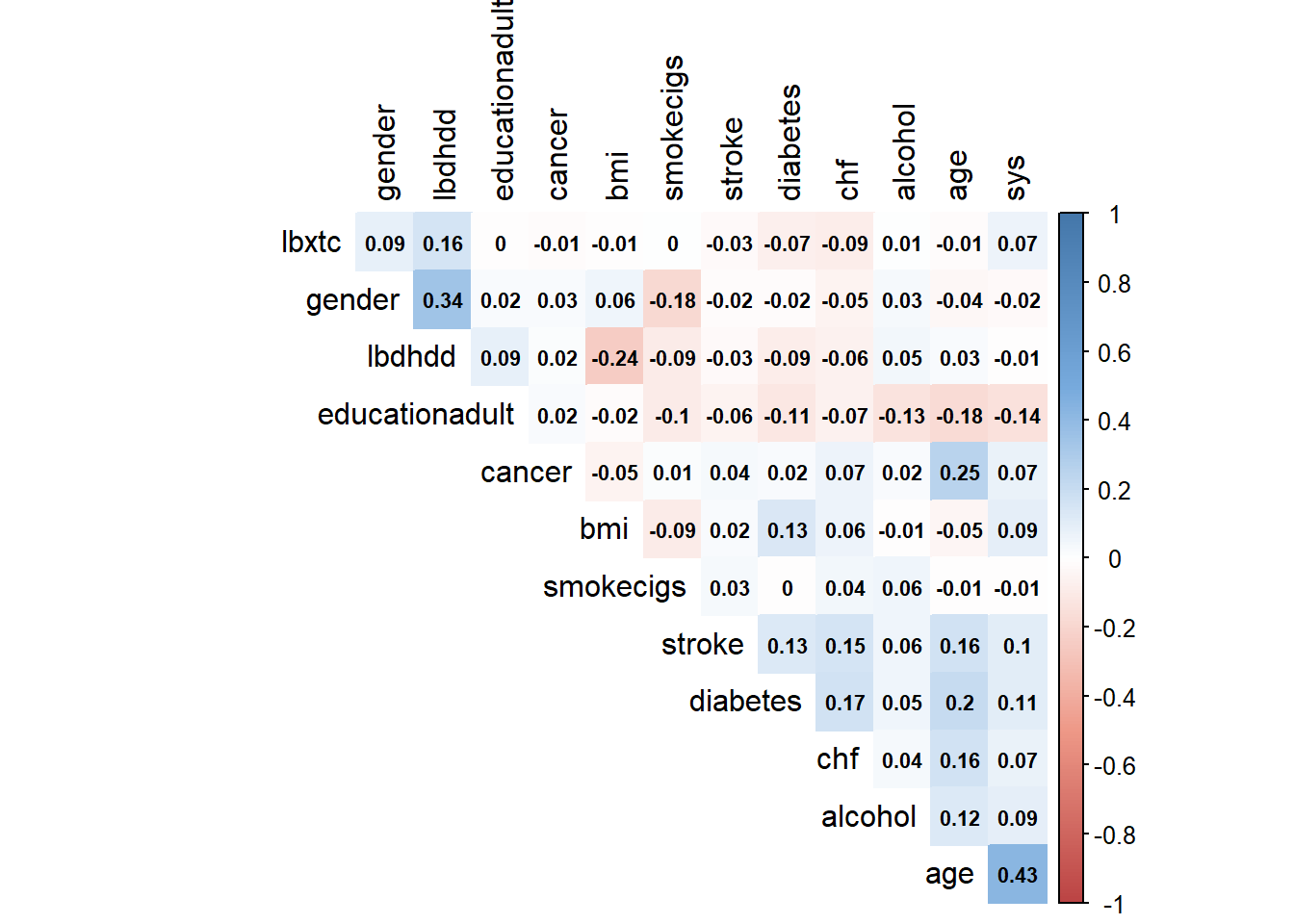
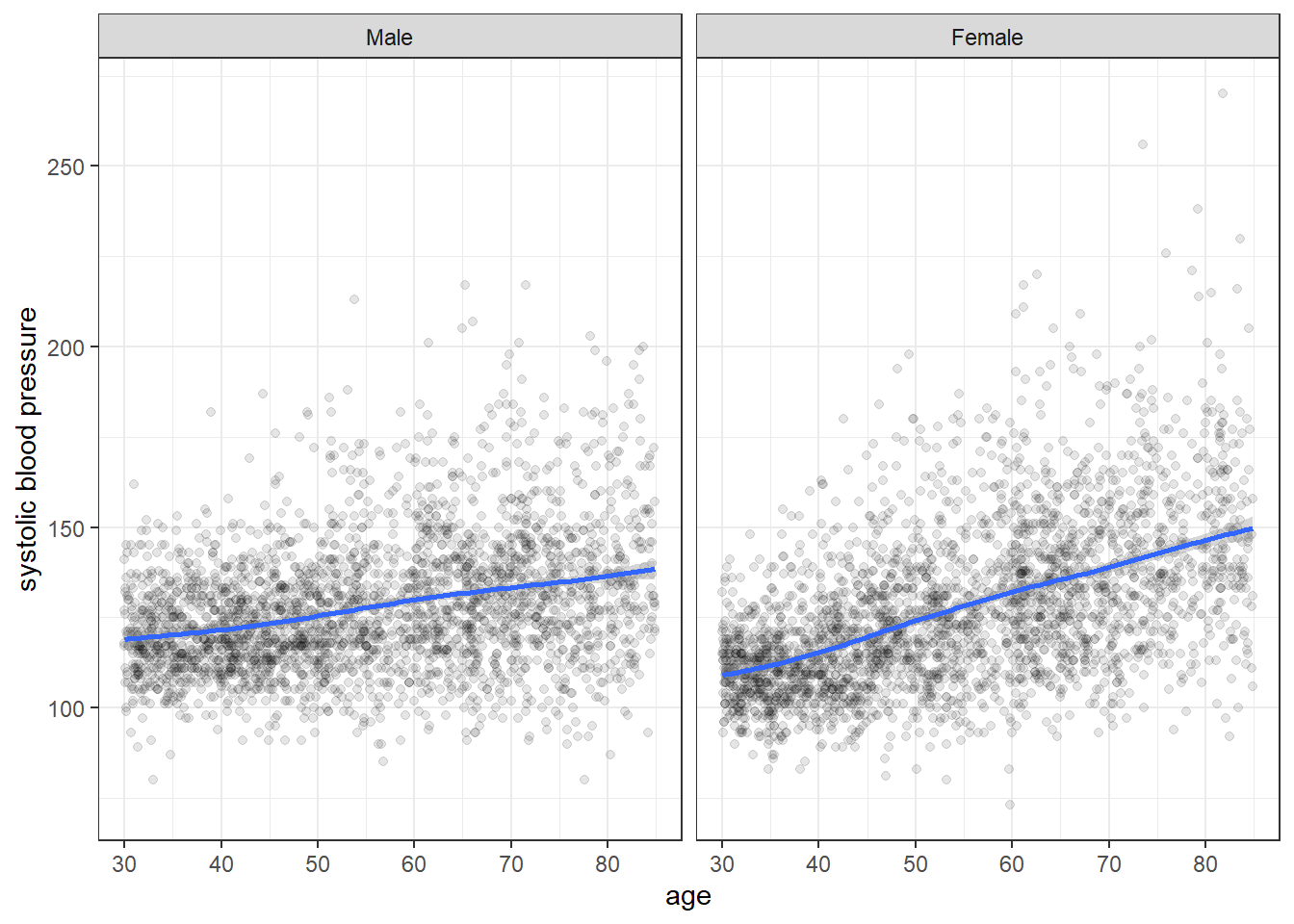
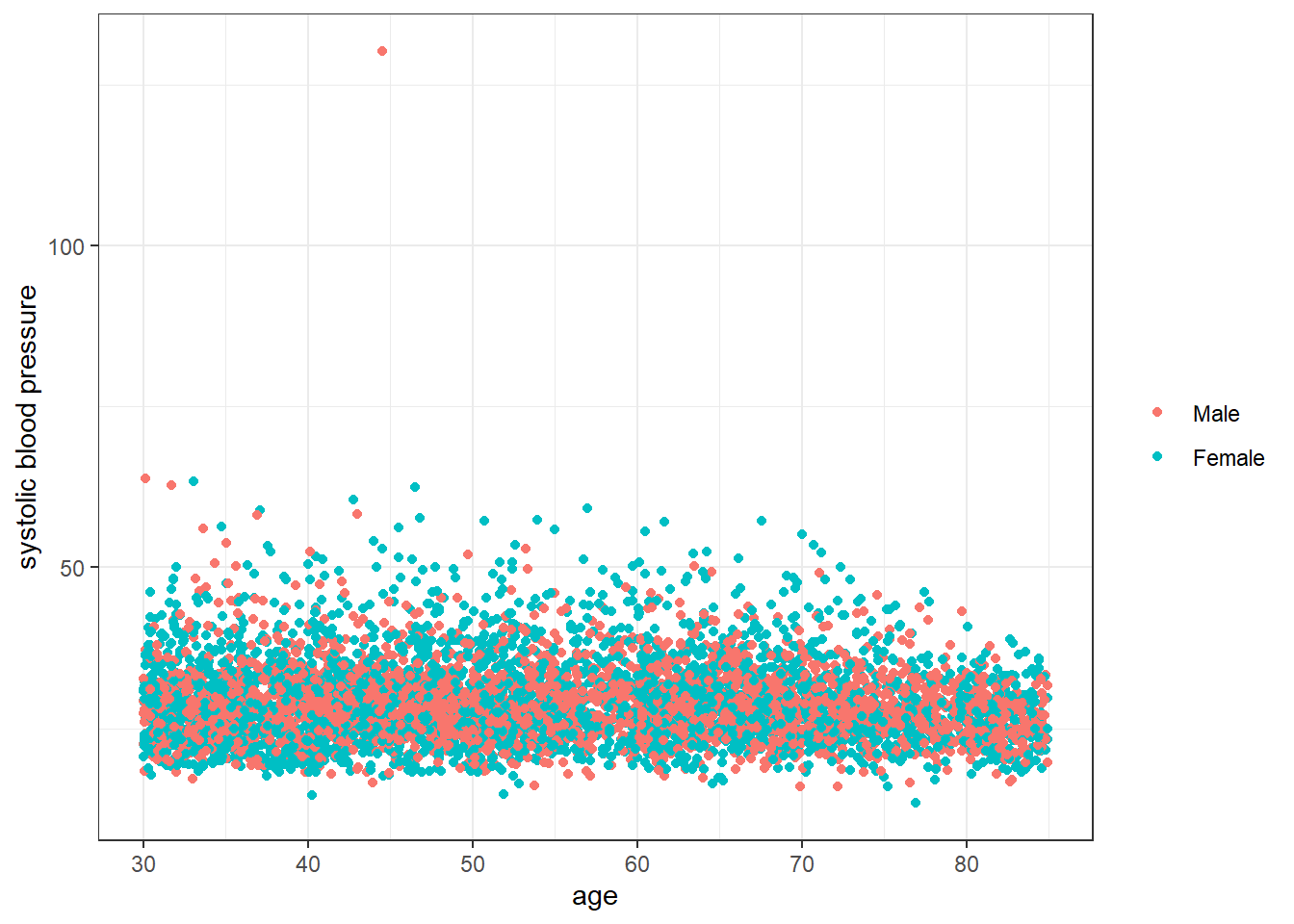
Below we further examine the association between age and systolic blood pressure stratified by pivotal variables.
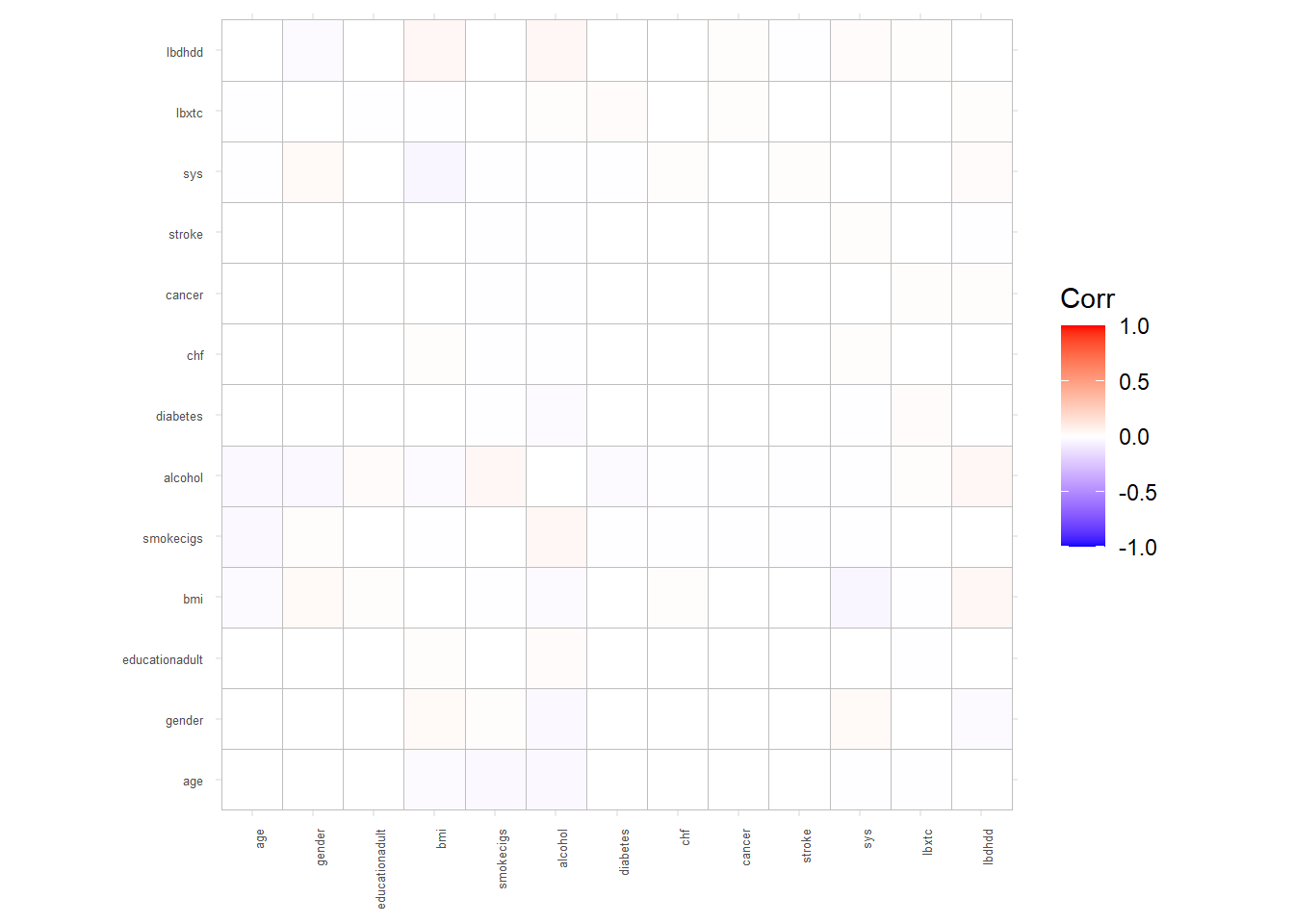
Next, we depict differences in the correlation coefficients (Spearman vs Pearson) in a heat map:
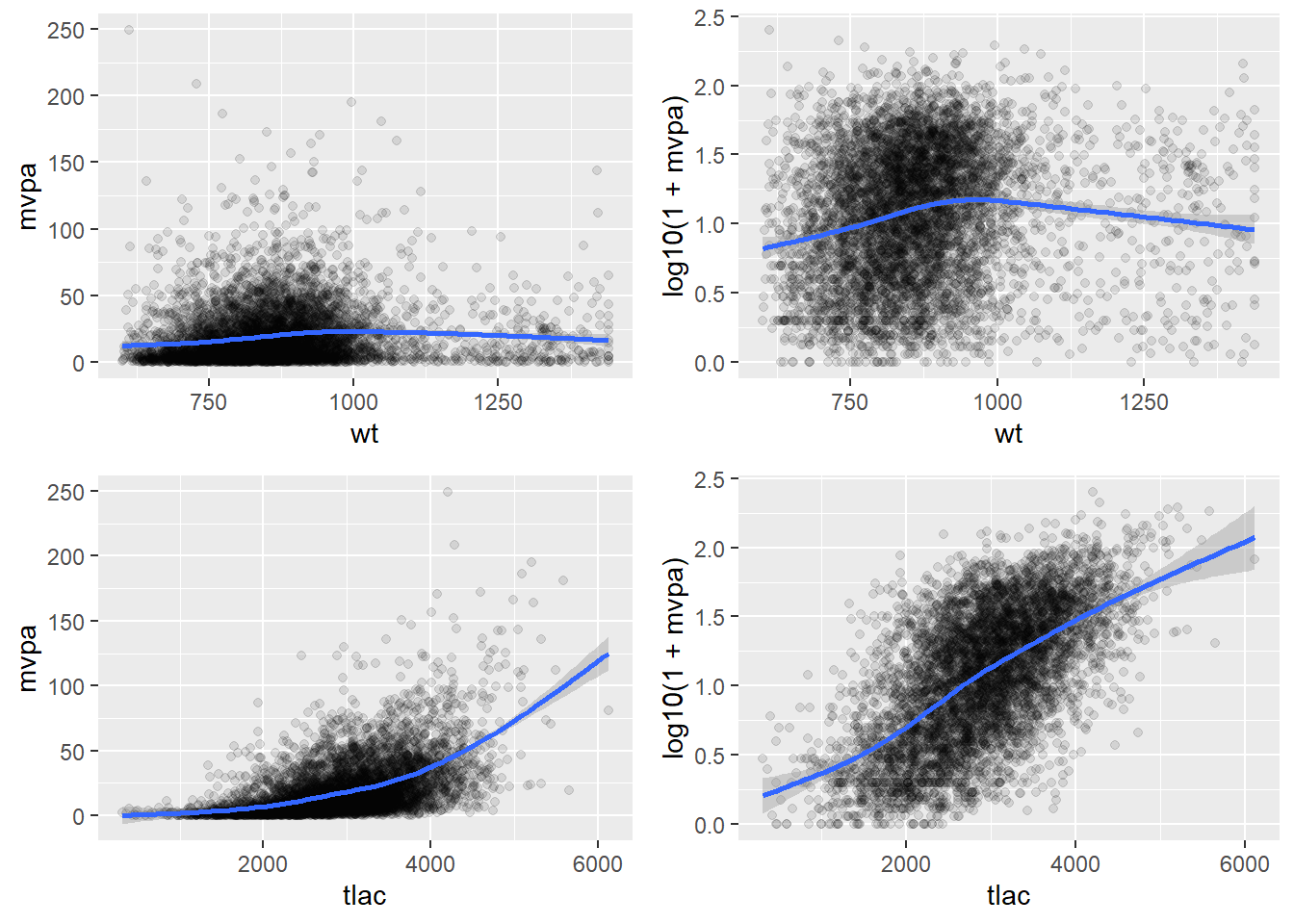
Correlations of the physical activity variables (outcome)
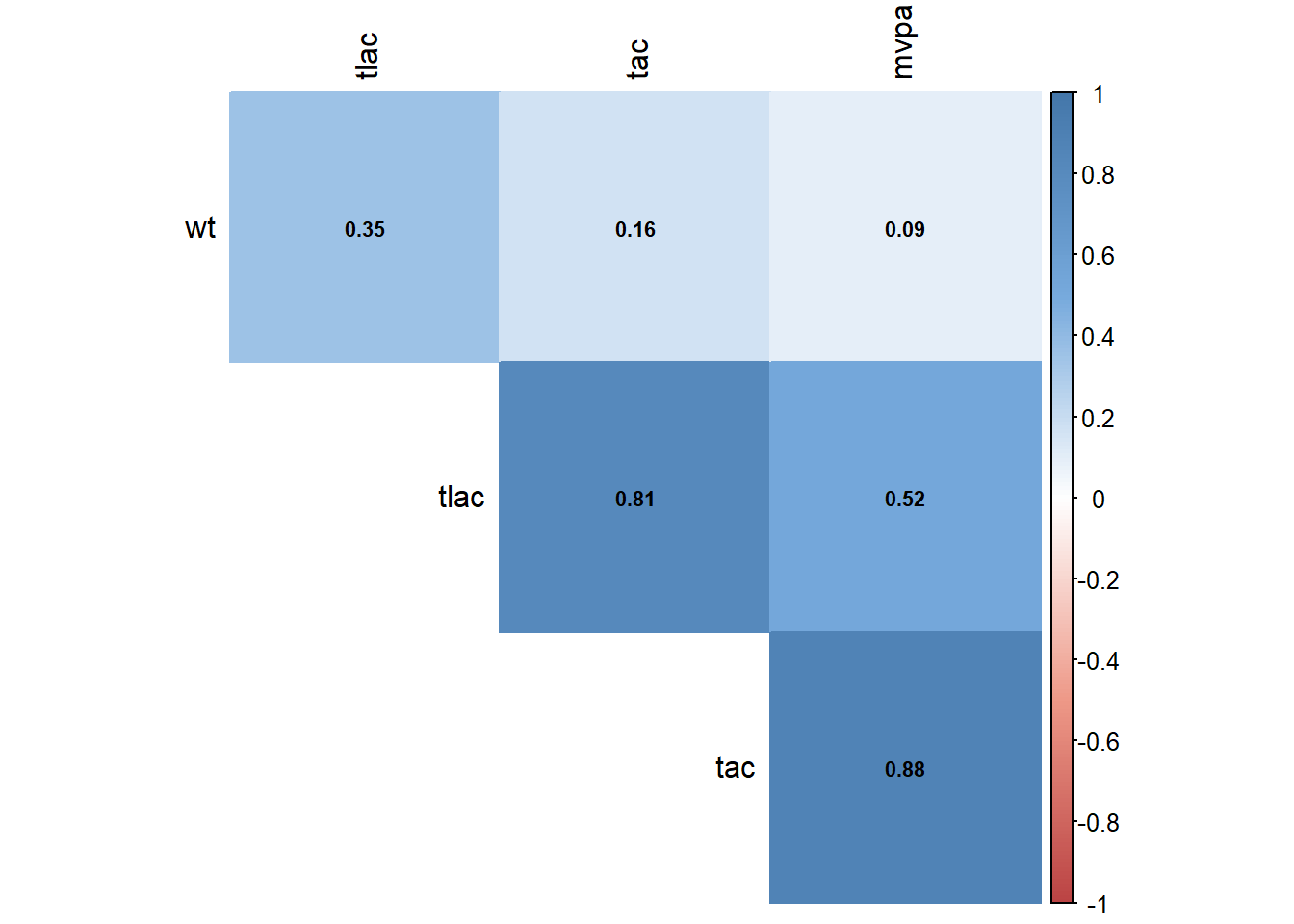
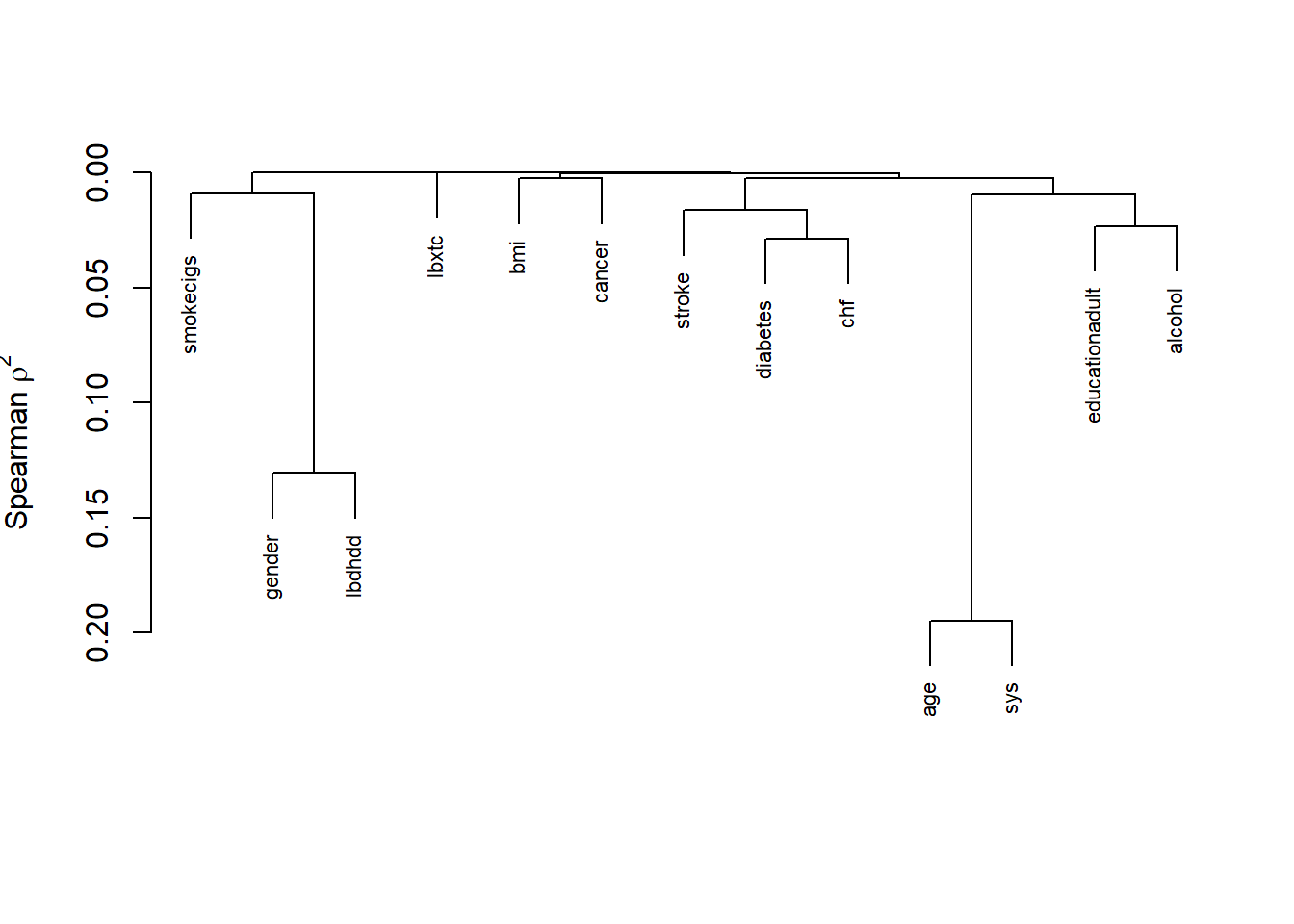
13.1.2 Variable clustering
Variable clustering is used for assessing collinearity, redundancy, and for separating variables into clusters that can be scored as a single variable, thus resulting in data reduction.
13.1.3 Variable redundancy
Redundancy analysis of predictor variables.
##
## Redundancy Analysis
##
## Hmisc::redun(formula = ~age + gender + bmi + sys + lbxtc + lbdhdd +
## smokecigs + diabetes + chf + cancer + stroke, data = a_nhanes)
##
## n: 5467 p: 11 nk: 3
##
## Number of NAs: 505
## Frequencies of Missing Values Due to Each Variable
## age gender bmi sys lbxtc lbdhdd smokecigs diabetes
## 0 0 44 274 230 230 2 0
## chf cancer stroke
## 0 0 0
##
##
## Transformation of target variables forced to be linear
##
## R-squared cutoff: 0.9 Type: ordinary
##
## R^2 with which each variable can be predicted from all other variables:
##
## age gender bmi sys lbxtc lbdhdd smokecigs diabetes
## 0.330 0.202 0.153 0.209 0.061 0.247 0.099 0.102
## chf cancer stroke
## 0.080 0.079 0.057
##
## No redundant variables13.3 Summary report by age group and gender
13.3.1 Summary report by gender
| Baseline characteristics by gender. | |||
| N |
Male N=2935 |
Female N=3037 |
|
|---|---|---|---|
age years |
5972 | 42.1 54.3 68.4 55.4 ± 15.3 |
41.2 53.0 66.2 54.3 ± 15.3 |
body mass index kg/m2 |
5928 | 24.99 27.85 31.24 28.59 ± 5.69 |
24.44 28.32 33.33 29.52 ± 7.03 |
| education level : Less than high school | 5968 | 0.29 856/2932 | 0.27 827/3036 |
| High school | 0.24 710/2932 | 0.24 738/3036 | |
| More than high school | 0.47 1366/2932 | 0.48 1471/3036 | |
Systolic blood pressure mg/dl |
5698 | 115.0 125.0 137.0 127.7 ± 18.3 |
111.0 123.0 139.0 127.1 ± 22.4 |
Total cholesterol mg/dl |
5742 | 172.0 198.0 224.0 200.4 ± 41.6 |
178.0 205.0 231.0 207.6 ± 42.7 |
HDL cholesterol mg/dl |
5742 | 40.0 46.0 56.0 49.0 ± 13.8 |
48.0 58.0 70.0 60.1 ± 17.1 |
| smoking status : Never | 5970 | 0.38 1119/2934 | 0.59 1792/3036 |
| Former | 0.36 1061/2934 | 0.23 698/3036 | |
| Current | 0.26 754/2934 | 0.18 546/3036 | |
| alcohol consumption : 1 | 5577 | 0.60 1645/2755 | 0.51 1445/2822 |
| 2 | 0.31 860/2755 | 0.44 1238/2822 | |
| 3 | 0.09 250/2755 | 0.05 139/2822 | |
| diabetes : Yes | 5972 | 0.13 383/2935 | 0.12 375/3037 |
| congestive heart failure : Yes | 5972 | 0.05 138/2935 | 0.03 95/3037 |
| cancer : Yes | 5972 | 0.10 280/2935 | 0.11 333/3037 |
| stroke : Yes | 5972 | 0.04 122/2935 | 0.04 116/3037 |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | |||
13.3.2 Summary report by age group for men
| Baseline characteristics by age group for men. | |||||
| N |
30-44 N=916 |
45-59 N=794 |
60-74 N=845 |
75+ N=380 |
|
|---|---|---|---|---|---|
body mass index kg/m2 |
2919 | 24.90 27.91 31.49 28.96 ± 6.82 |
25.18 27.96 31.31 28.65 ± 5.28 |
25.49 28.16 31.79 28.80 ± 5.11 |
24.20 26.65 29.72 27.14 ± 4.33 |
| education level : Less than high school | 2932 | 0.24 220/916 | 0.21 168/793 | 0.37 311/844 | 0.41 157/379 |
| High school | 0.26 236/916 | 0.25 201/793 | 0.23 194/844 | 0.21 79/379 | |
| More than high school | 0.50 460/916 | 0.53 424/793 | 0.40 339/844 | 0.38 143/379 | |
Systolic blood pressure mg/dl |
2824 | 112.0 119.0 129.0 120.9 ± 12.5 |
115.0 123.0 134.0 126.0 ± 17.1 |
120.0 131.0 145.0 133.3 ± 19.8 |
120.0 133.0 147.0 135.5 ± 22.2 |
Total cholesterol mg/dl |
2838 | 177.0 199.0 229.8 204.2 ± 42.6 |
178.0 204.0 230.0 206.3 ± 41.1 |
169.0 194.0 222.0 196.9 ± 41.0 |
159.0 185.0 211.8 187.2 ± 37.6 |
HDL cholesterol mg/dl |
2838 | 39.0 45.0 54.0 47.7 ± 13.6 |
40.0 47.0 57.0 49.5 ± 14.1 |
40.0 46.0 57.0 49.2 ± 13.5 |
41.0 47.0 57.0 50.7 ± 14.3 |
| smoking status : Never | 2934 | 0.50 454/916 | 0.39 308/794 | 0.28 234/845 | 0.32 123/379 |
| Former | 0.19 177/916 | 0.28 224/794 | 0.52 436/845 | 0.59 224/379 | |
| Current | 0.31 285/916 | 0.33 262/794 | 0.21 175/845 | 0.08 32/379 | |
| alcohol consumption : 1 | 2755 | 0.68 567/839 | 0.64 474/746 | 0.53 429/811 | 0.49 175/359 |
| 2 | 0.21 178/839 | 0.26 196/746 | 0.39 316/811 | 0.47 170/359 | |
| 3 | 0.11 94/839 | 0.10 76/746 | 0.08 66/811 | 0.04 14/359 | |
| diabetes : Yes | 2935 | 0.05 46/916 | 0.10 80/794 | 0.23 194/845 | 0.17 63/380 |
| congestive heart failure : Yes | 2935 | 0.01 5/916 | 0.03 23/794 | 0.08 69/845 | 0.11 41/380 |
| cancer : Yes | 2935 | 0.02 15/916 | 0.04 35/794 | 0.15 127/845 | 0.27 103/380 |
| stroke : Yes | 2935 | 0.00 4/916 | 0.02 14/794 | 0.07 59/845 | 0.12 45/380 |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | |||||
13.3.3 Summary report by age group for women
| Baseline characteristics by age group for women. | |||||
| N |
30-44 N=1006 |
45-59 N=838 |
60-74 N=844 |
75+ N=349 |
|
|---|---|---|---|---|---|
body mass index kg/m2 |
3009 | 24.06 28.11 33.33 29.29 ± 7.10 |
24.66 29.09 34.90 30.40 ± 7.58 |
24.92 28.76 33.36 29.81 ± 6.77 |
23.59 26.97 30.39 27.33 ± 5.36 |
| education level : Less than high school | 3036 | 0.21 215/1006 | 0.20 170/ 838 | 0.35 297/ 844 | 0.42 145/ 348 |
| High school | 0.20 200/1006 | 0.24 204/ 838 | 0.27 230/ 844 | 0.30 104/ 348 | |
| More than high school | 0.59 591/1006 | 0.55 464/ 838 | 0.38 317/ 844 | 0.28 99/ 348 | |
Systolic blood pressure mg/dl |
2874 | 104.0 111.0 120.0 113.1 ± 13.2 |
113.0 124.0 136.0 125.9 ± 19.4 |
121.0 134.0 150.0 137.0 ± 22.2 |
131.2 142.5 158.8 146.0 ± 24.7 |
Total cholesterol mg/dl |
2904 | 171.0 196.0 223.8 199.8 ± 42.8 |
181.0 207.0 231.5 209.6 ± 41.9 |
186.0 212.0 239.0 214.9 ± 42.7 |
179.0 204.0 232.0 207.6 ± 41.0 |
HDL cholesterol mg/dl |
2904 | 47.0 57.0 69.0 59.6 ± 17.2 |
47.0 58.0 70.0 60.3 ± 17.4 |
49.0 58.0 69.0 59.8 ± 16.4 |
49.0 60.0 73.8 62.1 ± 17.4 |
| smoking status : Never | 3036 | 0.65 655/1006 | 0.55 459/ 837 | 0.55 466/ 844 | 0.61 212/ 349 |
| Former | 0.14 137/1006 | 0.23 191/ 837 | 0.30 255/ 844 | 0.33 115/ 349 | |
| Current | 0.21 214/1006 | 0.22 187/ 837 | 0.15 123/ 844 | 0.06 22/ 349 | |
| alcohol consumption : 1 | 2822 | 0.63 568/908 | 0.56 433/776 | 0.41 332/811 | 0.34 112/327 |
| 2 | 0.33 299/908 | 0.36 283/776 | 0.55 450/811 | 0.63 206/327 | |
| 3 | 0.05 41/908 | 0.08 60/776 | 0.04 29/811 | 0.03 9/327 | |
| diabetes : Yes | 3037 | 0.03 30/1006 | 0.12 104/ 838 | 0.21 178/ 844 | 0.18 63/ 349 |
| congestive heart failure : Yes | 3037 | 0.01 8/1006 | 0.02 16/ 838 | 0.05 40/ 844 | 0.09 31/ 349 |
| cancer : Yes | 3037 | 0.04 36/1006 | 0.10 84/ 838 | 0.14 121/ 844 | 0.26 92/ 349 |
| stroke : Yes | 3037 | 0.01 12/1006 | 0.03 24/ 838 | 0.06 47/ 844 | 0.09 33/ 349 |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | |||||
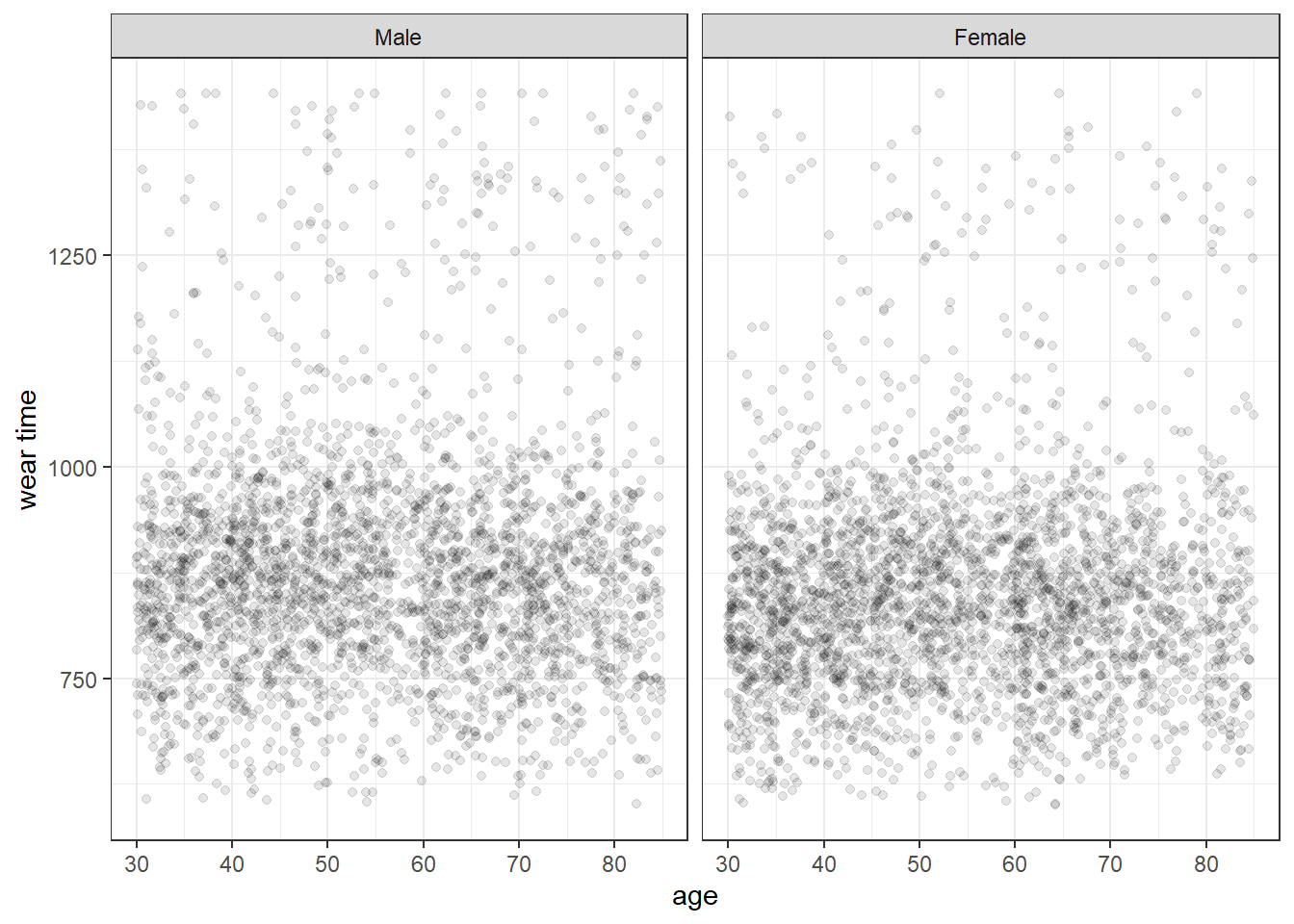
13.4 Continuous variables by age and gender
13.6 Section session info
## R version 4.1.3 (2022-03-10)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_Austria.1252 LC_CTYPE=English_Austria.1252
## [3] LC_MONETARY=English_Austria.1252 LC_NUMERIC=C
## [5] LC_TIME=English_Austria.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] gridExtra_2.3 naniar_0.6.1 ggcorrplot_0.1.3 corrplot_0.92
## [5] gtsummary_1.5.2 Hmisc_4.6-0 Formula_1.2-4 survival_3.2-13
## [9] lattice_0.20-45 plotly_4.10.0 forcats_0.5.1 stringr_1.4.0
## [13] dplyr_1.0.8 purrr_0.3.4 readr_2.1.2 tidyr_1.2.0
## [17] tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1 here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-155 fs_1.5.2 lubridate_1.8.0
## [4] RColorBrewer_1.1-2 httr_1.4.2 rprojroot_2.0.2
## [7] tools_4.1.3 backports_1.4.1 bslib_0.3.1
## [10] utf8_1.2.2 R6_2.5.1 rpart_4.1.16
## [13] mgcv_1.8-39 DBI_1.1.2 lazyeval_0.2.2
## [16] colorspace_2.0-3 nnet_7.3-17 withr_2.5.0
## [19] tidyselect_1.1.2 compiler_4.1.3 cli_3.2.0
## [22] rvest_1.0.2 gt_0.4.0 htmlTable_2.4.0
## [25] xml2_1.3.3 labeling_0.4.2 bookdown_0.25
## [28] sass_0.4.1 checkmate_2.0.0 scales_1.1.1
## [31] digest_0.6.29 foreign_0.8-82 rmarkdown_2.13
## [34] base64enc_0.1-3 jpeg_0.1-9 pkgconfig_2.0.3
## [37] htmltools_0.5.2 highr_0.9 dbplyr_2.1.1
## [40] fastmap_1.1.0 htmlwidgets_1.5.4 rlang_1.0.2
## [43] readxl_1.3.1 rstudioapi_0.13 farver_2.1.0
## [46] jquerylib_0.1.4 generics_0.1.2 jsonlite_1.8.0
## [49] crosstalk_1.2.0 magrittr_2.0.2 Matrix_1.4-0
## [52] Rcpp_1.0.8.3 munsell_0.5.0 fansi_1.0.3
## [55] visdat_0.5.3 lifecycle_1.0.1 stringi_1.7.6
## [58] yaml_2.3.5 plyr_1.8.7 grid_4.1.3
## [61] crayon_1.5.1 haven_2.4.3 splines_4.1.3
## [64] hms_1.1.1 knitr_1.38 pillar_1.7.0
## [67] reshape2_1.4.4 reprex_2.0.1 glue_1.6.2
## [70] evaluate_0.15 latticeExtra_0.6-29 broom.helpers_1.6.0
## [73] data.table_1.14.2 modelr_0.1.8 vctrs_0.3.8
## [76] png_0.1-7 tzdb_0.2.0 cellranger_1.1.0
## [79] gtable_0.3.0 assertthat_0.2.1 xfun_0.30
## [82] broom_0.7.12 viridisLite_0.4.0 cluster_2.1.2
## [85] ellipsis_0.3.2