Chapter 8 Multivariate distributions
8.1 Overview
8.1.1 Variable correlation
corrs <- a_crash2 %>%
dplyr::select(age, sex, sbp, hr, rr ,cc, injurytime, injurytype ) %>%
filter(complete.cases(.)) %>%
dplyr::mutate_all(as.numeric)
M <- cor(corrs)
col <- colorRampPalette(c("#BB4444", "#EE9988", "#FFFFFF", "#77AADD", "#4477AA"))
corrplot(M, method = "color", col = col(200),
type = "upper", order = "hclust", number.cex = .7,
addCoef.col = "black", # Add coefficient of correlation
tl.col = "black", tl.srt = 90, # Text label color and rotation
# hide correlation coefficient on the principal diagonal
diag = FALSE)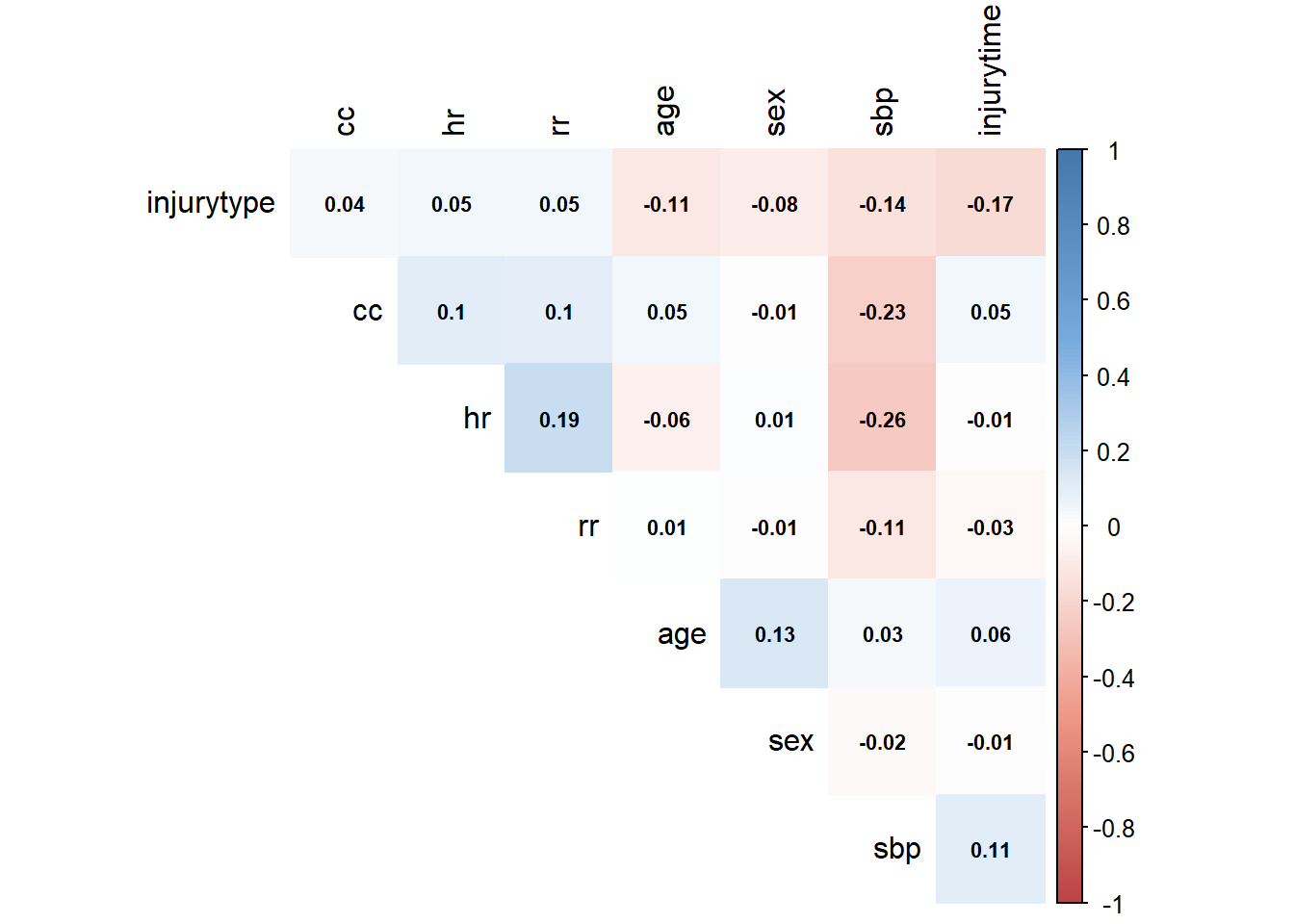
8.1.2 Variable clustering
Variable clustering is used for assessing collinearity, redundancy, and for separating variables into clusters that can be scored as a single variable, thus resulting in data reduction.
Hmisc::varclus( ~ age + sbp + hr + rr + cc + gcs + injurytime + injurytype + sex, data = a_crash2)## Hmisc::varclus(x = ~age + sbp + hr + rr + cc + gcs + injurytime +
## injurytype + sex, data = a_crash2)
##
##
## Similarity matrix (Spearman rho^2)
##
## age sbp hr rr cc gcs injurytime
## age 1.00 0.00 0.00 0.00 0.00 0.00 0.01
## sbp 0.00 1.00 0.11 0.03 0.07 0.01 0.01
## hr 0.00 0.11 1.00 0.05 0.02 0.02 0.00
## rr 0.00 0.03 0.05 1.00 0.02 0.00 0.00
## cc 0.00 0.07 0.02 0.02 1.00 0.02 0.00
## gcs 0.00 0.01 0.02 0.00 0.02 1.00 0.01
## injurytime 0.01 0.01 0.00 0.00 0.00 0.01 1.00
## injurytypepenetrating 0.02 0.00 0.01 0.00 0.00 0.06 0.05
## injurytypeblunt and penetrating 0.00 0.01 0.01 0.00 0.00 0.01 0.00
## sexfemale 0.01 0.00 0.00 0.00 0.00 0.00 0.00
## injurytypepenetrating
## age 0.02
## sbp 0.00
## hr 0.01
## rr 0.00
## cc 0.00
## gcs 0.06
## injurytime 0.05
## injurytypepenetrating 1.00
## injurytypeblunt and penetrating 0.07
## sexfemale 0.02
## injurytypeblunt and penetrating sexfemale
## age 0.00 0.01
## sbp 0.01 0.00
## hr 0.01 0.00
## rr 0.00 0.00
## cc 0.00 0.00
## gcs 0.01 0.00
## injurytime 0.00 0.00
## injurytypepenetrating 0.07 0.02
## injurytypeblunt and penetrating 1.00 0.00
## sexfemale 0.00 1.00
##
## No. of observations used for each pair:
##
## age sbp hr rr cc gcs injurytime
## age 20203 19884 20066 20012 19593 20180 20193
## sbp 19884 19887 19795 19750 19316 19883 19877
## hr 20066 19795 20070 19943 19482 20066 20059
## rr 20012 19750 19943 20016 19454 20014 20008
## cc 19593 19316 19482 19454 19596 19595 19588
## gcs 20180 19883 20066 20014 19595 20184 20173
## injurytime 20193 19877 20059 20008 19588 20173 20196
## injurytypepenetrating 20203 19887 20070 20016 19596 20184 20196
## injurytypeblunt and penetrating 20203 19887 20070 20016 19596 20184 20196
## sexfemale 20202 19886 20069 20015 19595 20183 20195
## injurytypepenetrating
## age 20203
## sbp 19887
## hr 20070
## rr 20016
## cc 19596
## gcs 20184
## injurytime 20196
## injurytypepenetrating 20207
## injurytypeblunt and penetrating 20207
## sexfemale 20206
## injurytypeblunt and penetrating sexfemale
## age 20203 20202
## sbp 19887 19886
## hr 20070 20069
## rr 20016 20015
## cc 19596 19595
## gcs 20184 20183
## injurytime 20196 20195
## injurytypepenetrating 20207 20206
## injurytypeblunt and penetrating 20207 20206
## sexfemale 20206 20206
##
## hclust results (method=complete)
##
##
## Call:
## hclust(d = as.dist(1 - x), method = method)
##
## Cluster method : complete
## Number of objects: 10Plot associations.
plot(Hmisc::varclus( ~ age + sbp + hr + rr + cc + gcs + injurytime + injurytype + sex, data = a_crash2))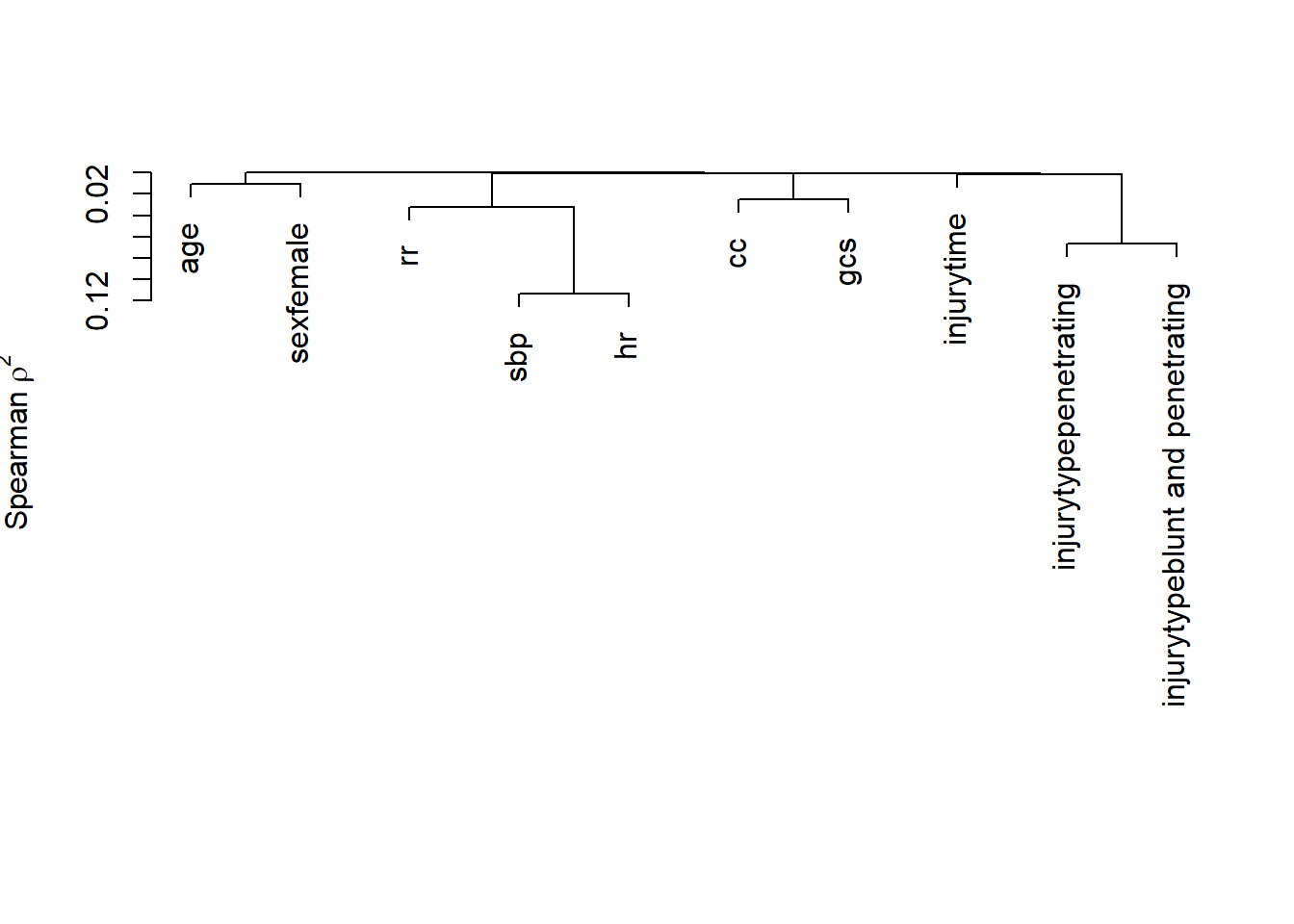
8.1.3 Variable redundancy
Redundancy analysis of predictor variables.
Hmisc::redun( ~ hr + rr + age + sbp + injurytype + sex , data = a_crash2)##
## Redundancy Analysis
##
## Hmisc::redun(formula = ~hr + rr + age + sbp + injurytype + sex,
## data = a_crash2)
##
## n: 19689 p: 6 nk: 3
##
## Number of NAs: 518
## Frequencies of Missing Values Due to Each Variable
## hr rr age sbp injurytype sex
## 137 191 4 320 0 1
##
##
## Transformation of target variables forced to be linear
##
## R-squared cutoff: 0.9 Type: ordinary
##
## R^2 with which each variable can be predicted from all other variables:
##
## hr rr age sbp injurytype sex
## 0.116 0.044 0.052 0.099 0.061 0.035
##
## No redundant variables8.2 Summary reports by sex
8.2.1 Overall
| Baseline characteristics by sex. | |||
| N |
male N=16935 |
female N=3271 |
|
|---|---|---|---|
Age years |
20203 | 23.0 30.0 41.0 33.7 ± 13.6 |
25.0 35.0 50.0 38.8 ± 16.8 |
Systolic Blood Pressure mmHg |
19887 | 80.0 95.0 110.0 98.8 ± 25.5 |
80.0 90.0 110.0 96.7 ± 25.7 |
Heart Rate /min |
20070 | 90.0 105.0 120.0 104.3 ± 21.2 |
92.0 106.0 120.0 105.2 ± 21.0 |
Respiratory Rate /min |
20016 | 20.00 22.00 26.00 23.07 ± 6.77 |
20.00 22.00 26.00 23.03 ± 6.58 |
Central Capillary Refille Time s |
19596 | 2.00 3.00 4.00 3.27 ± 1.72 |
2.00 3.00 4.00 3.23 ± 1.59 |
Glasgow Coma Score Total points |
20184 | 11.00 15.00 15.00 12.44 ± 3.72 |
12.00 14.00 15.00 12.62 ± 3.46 |
Hours Since Injury hours |
20196 | 1.00 2.00 4.00 2.85 ± 2.39 |
1.00 2.00 4.00 2.84 ± 2.67 |
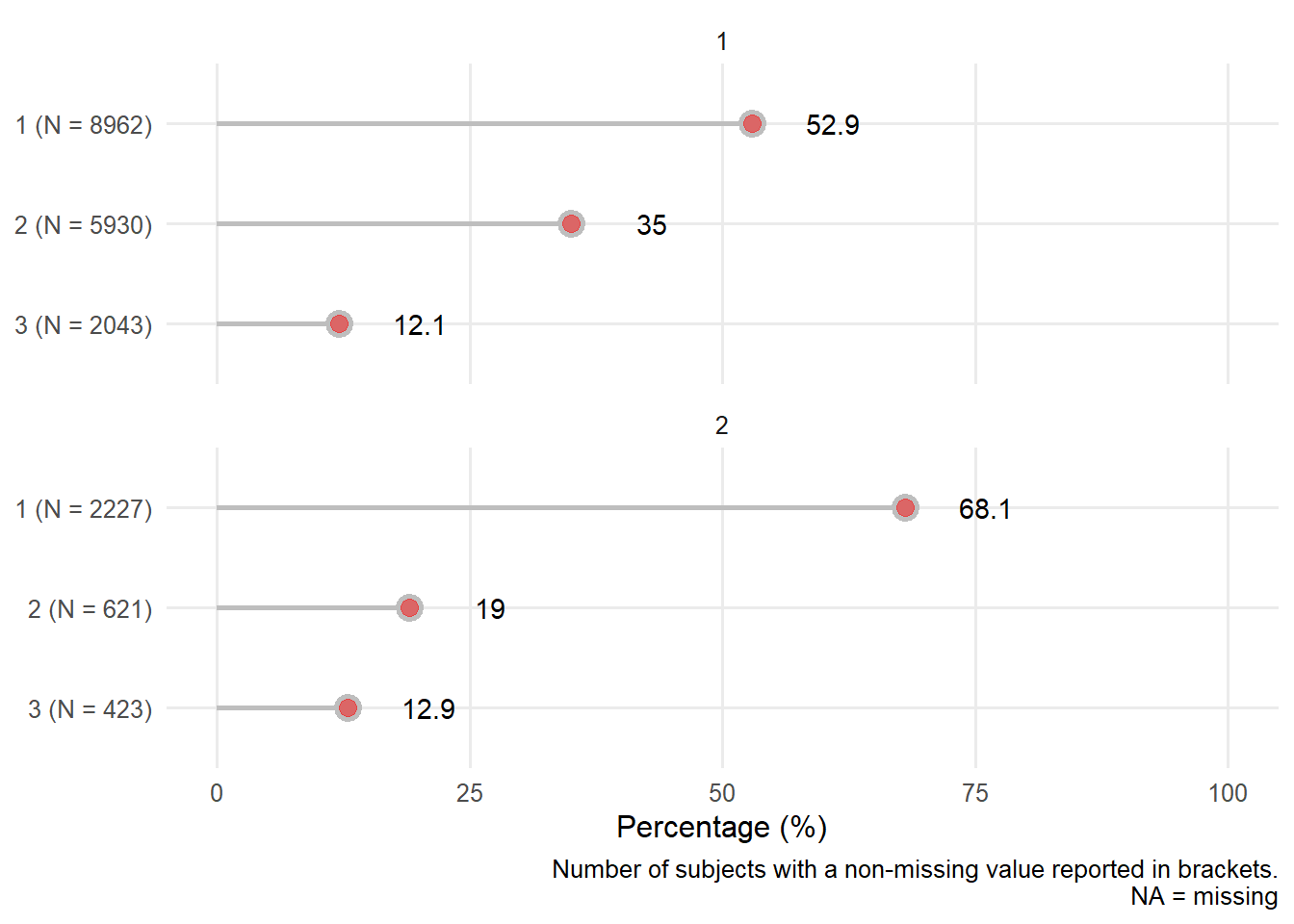
| Injury type : blunt | 20207 | 0.53 8962/16935 | 0.68 2227/ 3271 |
| penetrating | 0.35 5930/16935 | 0.19 621/ 3271 | |
| blunt and penetrating | 0.12 2043/16935 | 0.13 423/ 3271 | |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | |||
8.2.3 Distribution of systolic blood pressure by sex
Figure 7.5: Distribution of systolic blood pressure by sex
8.2.6 Distribution of central capillary refille time by sex
Figure 8.2: Distribution of central capillary refill time by sex
8.2.7 Distribution of hours since injury by sex
Figure 8.3: Distribution of hours since injury by sex
8.3 Summary reports by age
Categorize age for the purposes of exploring the relationship between age and other baseline variables. This is purely for exploratory purposes only, and not to influence the analysis strategy by pursuing the dichotomization of age.
| Characteristic | N = 20,2071 |
|---|---|
| age_C | |
| <30 | 9,070 (45%) |
| 30-44 | 6,477 (32%) |
| 45-59 | 3,204 (16%) |
| 60+ | 1,452 (7.2%) |
| NA | 4 (<0.1%) |
| 1 n (%) | |
Report all variables by age category.
| Baseline characteristics by age categories. | |||||
| N |
<30 N=9070 |
30-44 N=6477 |
45-59 N=3204 |
60+ N=1452 |
|
|---|---|---|---|---|---|
| Sex : female | 20202 | 0.13 1183/9070 | 0.15 959/6476 | 0.21 659/3204 | 0.32 469/1452 |
Systolic Blood Pressure mmHg |
19884 | 80.0 96.0 110.0 98.1 ± 23.8 |
80.0 90.0 110.0 97.7 ± 25.3 |
80.0 94.0 112.0 100.1 ± 28.4 |
80.0 90.0 110.0 100.4 ± 30.2 |
Heart Rate /min |
20066 | 91.0 106.0 120.0 105.3 ± 21.3 |
90.0 106.0 120.0 104.7 ± 20.9 |
90.0 104.0 120.0 103.3 ± 21.0 |
88.0 100.0 116.0 101.0 ± 21.8 |
Respiratory Rate /min |
20012 | 20.00 22.00 26.00 22.93 ± 6.74 |
20.00 22.00 26.00 23.24 ± 6.68 |
20.00 22.00 26.00 23.11 ± 6.80 |
20.00 22.00 26.00 23.04 ± 6.89 |
Central Capillary Refille Time s |
19593 | 2.00 3.00 4.00 3.20 ± 1.77 |
2.00 3.00 4.00 3.27 ± 1.65 |
2.00 3.00 4.00 3.34 ± 1.64 |
2.00 3.00 4.00 3.48 ± 1.56 |
Glasgow Coma Score Total points |
20180 | 11.00 15.00 15.00 12.64 ± 3.61 |
11.00 14.50 15.00 12.39 ± 3.72 |
11.00 14.00 15.00 12.38 ± 3.70 |
10.00 14.00 15.00 12.00 ± 3.82 |
Hours Since Injury hours |
20193 | 1.00 2.00 4.00 2.71 ± 2.18 |
1.00 2.00 4.00 2.83 ± 2.28 |
1.00 2.50 4.50 3.12 ± 3.17 |
1.00 3.00 4.50 3.12 ± 2.68 |
| Injury type : blunt | 20203 | 0.50 4544/9070 | 0.53 3462/6477 | 0.65 2081/3204 | 0.76 1101/1452 |
| penetrating | 0.38 3448/9070 | 0.33 2155/6477 | 0.23 748/3204 | 0.14 199/1452 | |
| blunt and penetrating | 0.12 1078/9070 | 0.13 860/6477 | 0.12 375/3204 | 0.10 152/1452 | |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | |||||
8.3.1 Distribution of systolic blood pressure by age categories
Figure 8.6: Distribution of systolic blood pressure by gcs
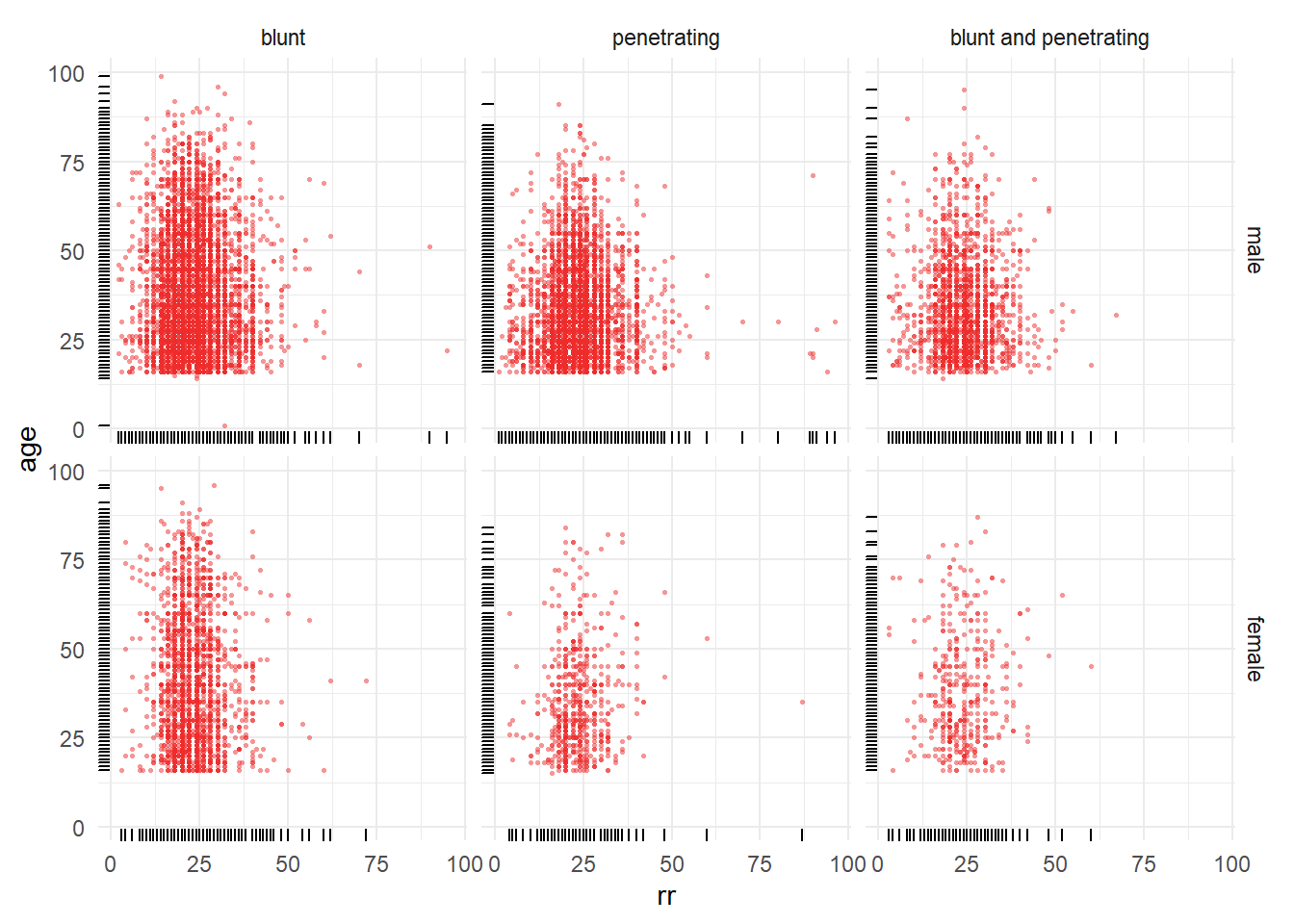
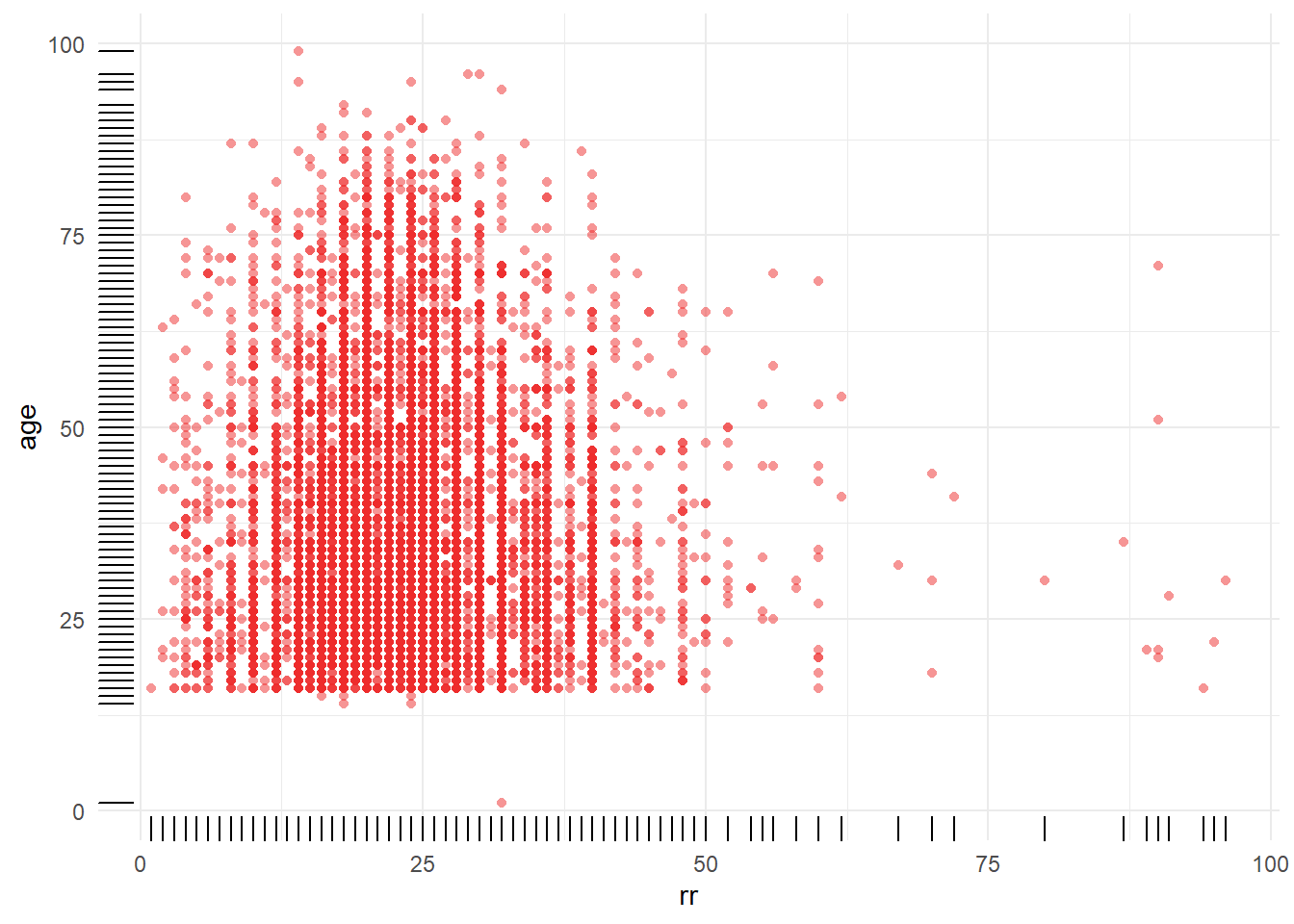
8.3.3 Distribution of respiratory rate by age categories
Figure 8.8: Distribution of respiratory rate by gcs
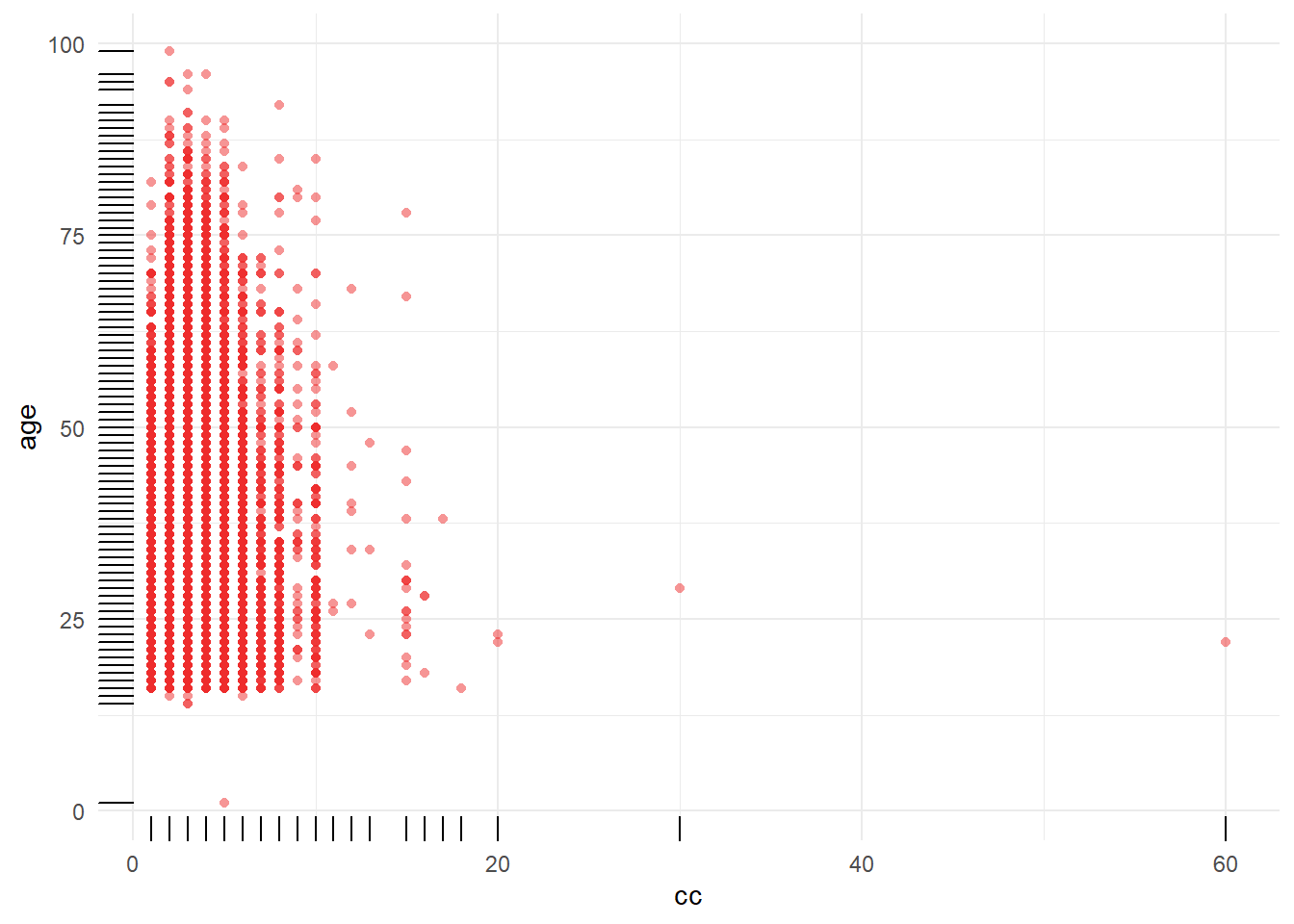
8.3.4 Distribution of central capillary refille time by age categories
Figure 8.9: Distribution of central capillary refill time by gcs
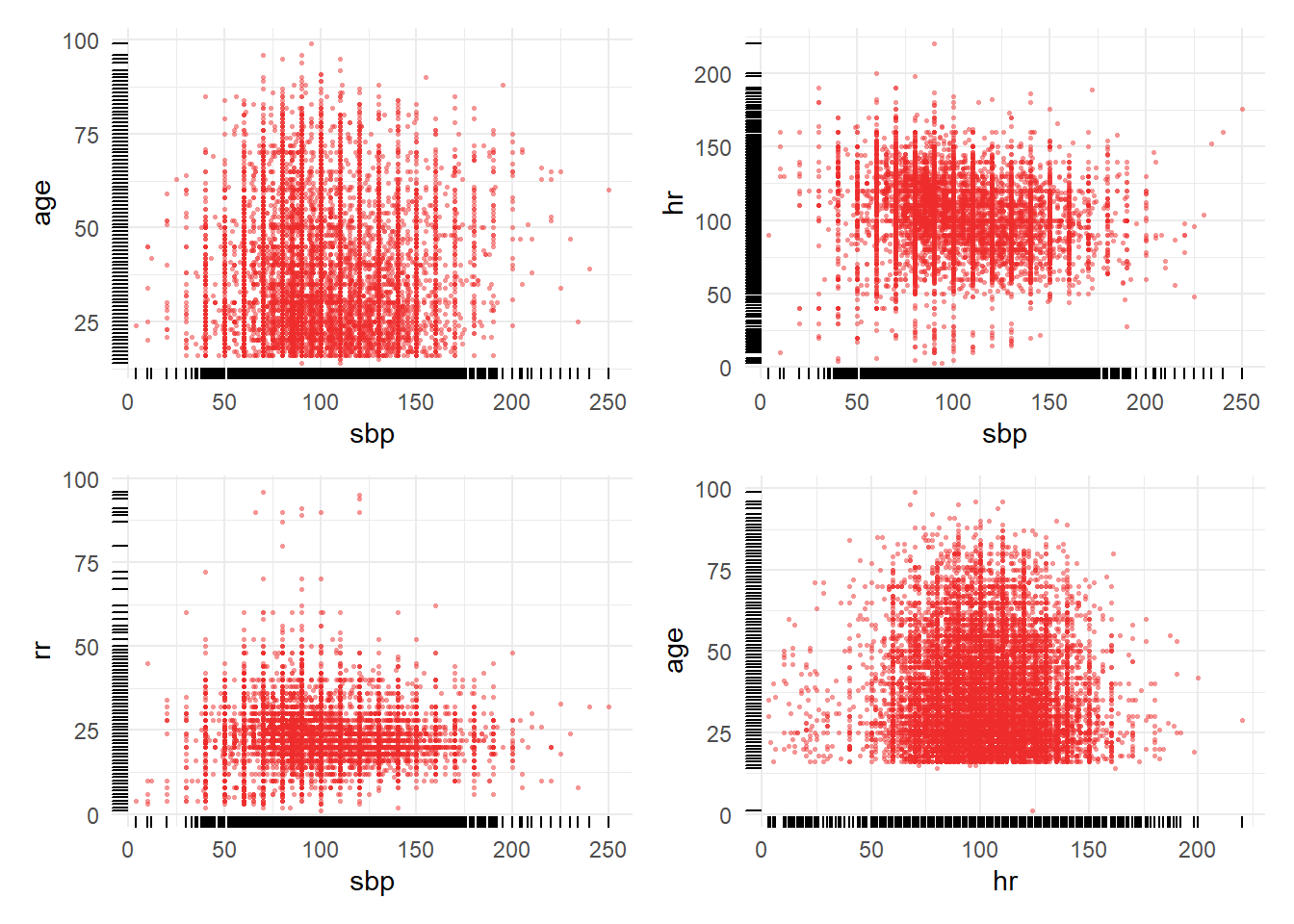
8.3.5 WIP: multivariate scatter plots
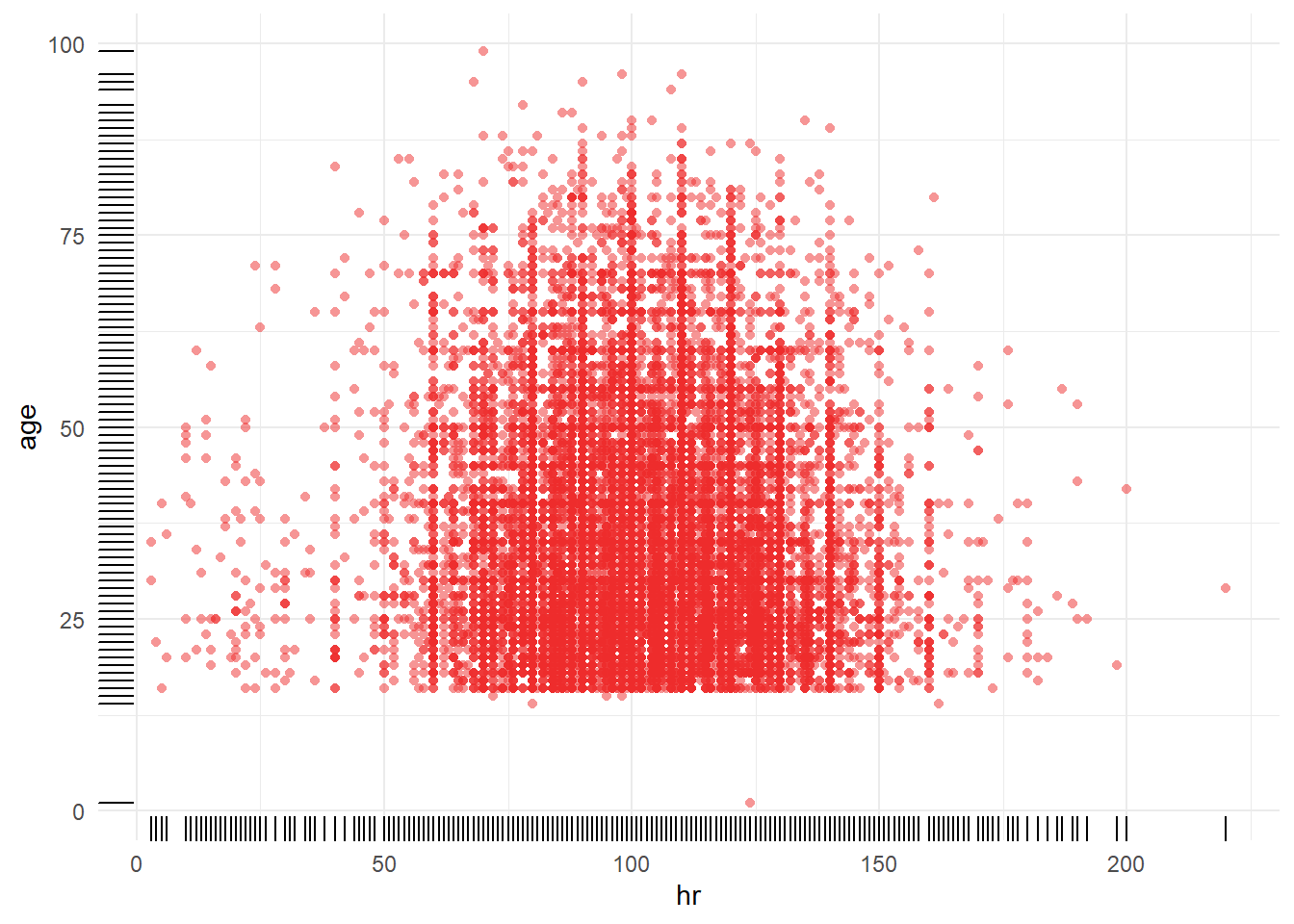
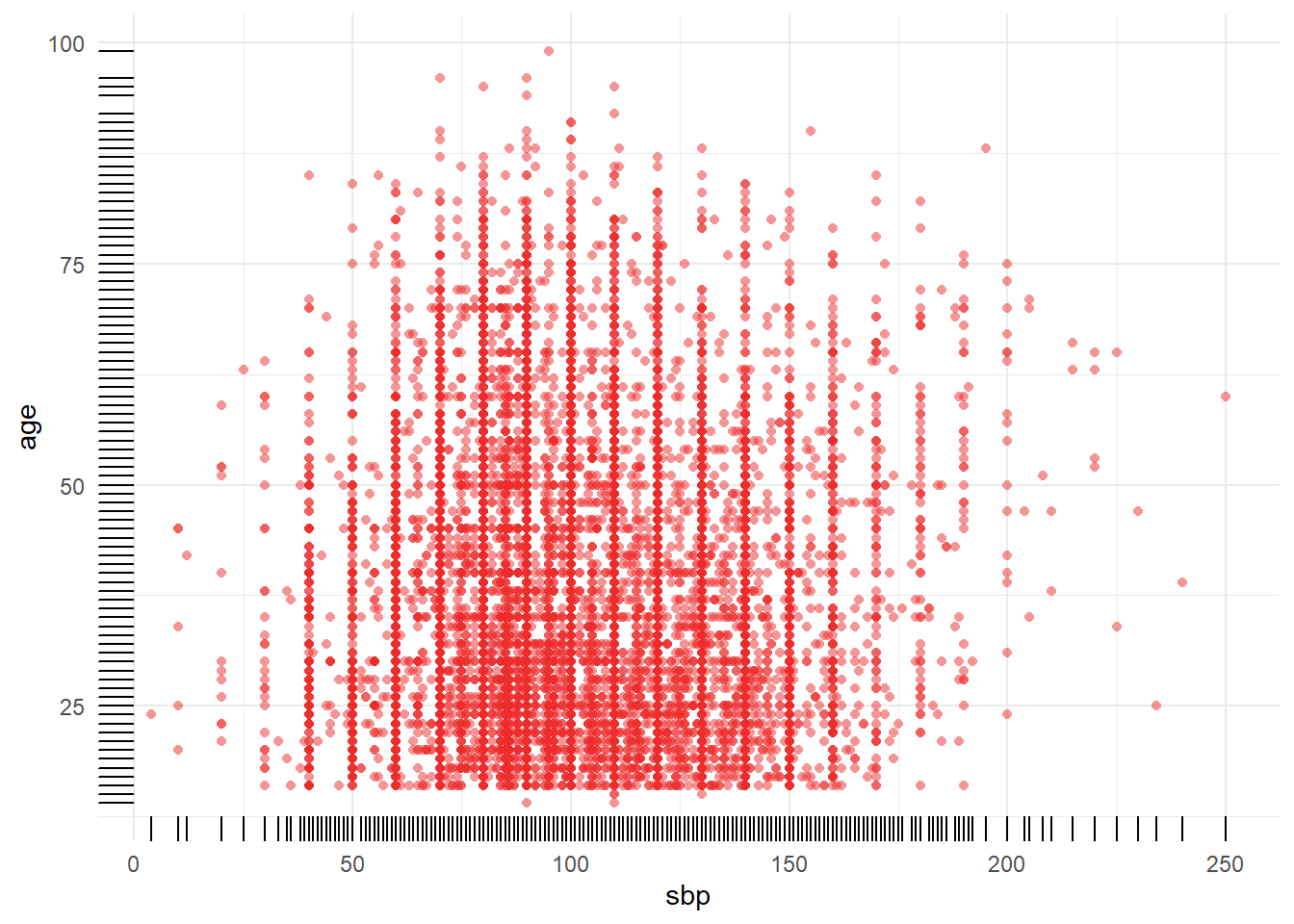
a_crash2 %>% dplyr::filter(!is.na(sbp)) %>% tally()## n
## 1 19887a_crash2 %>% dplyr::filter(is.na(sbp)) %>% tally()## n
## 1 320bigN <- a_crash2 %>% dplyr::filter(!is.na(sbp) & !is.na(age)) %>% tally()
n_miss <- a_crash2 %>% dplyr::filter(is.na(sbp) | is.na(age)) %>% tally()
title <-
paste0("Plot of ", Hmisc::label(a_crash2$age), " and ", Hmisc::label(a_crash2$sbp))
caption <-
paste0(
"n = ",
bigN,
" subjects displayed.\n",
n_miss,
" subjects with a missing value in at least one of the variables."
)
x_axis <- paste0(Hmisc::label(a_crash2$age), " [", Hmisc::units(a_crash2$age), "]")
y_axis <- paste0(Hmisc::label(a_crash2$sbp), " [", Hmisc::units(a_crash2$sbp), "]")
p1 <- a_crash2 %>%
dplyr::filter(!is.na(sbp) & !is.na(age)) %>%
mutate(sbp = as.numeric(sbp),
age = as.numeric(age)) %>%
ggplot(aes(x = sbp, y = age)) +
ylab(x_axis) +
xlab(y_axis) +
labs(
title = title,
caption = caption
) +
geom_point(shape = 16, #size = 0.5,
alpha = 0.5,
color = "firebrick2") +
geom_rug() +
theme_minimal()
p1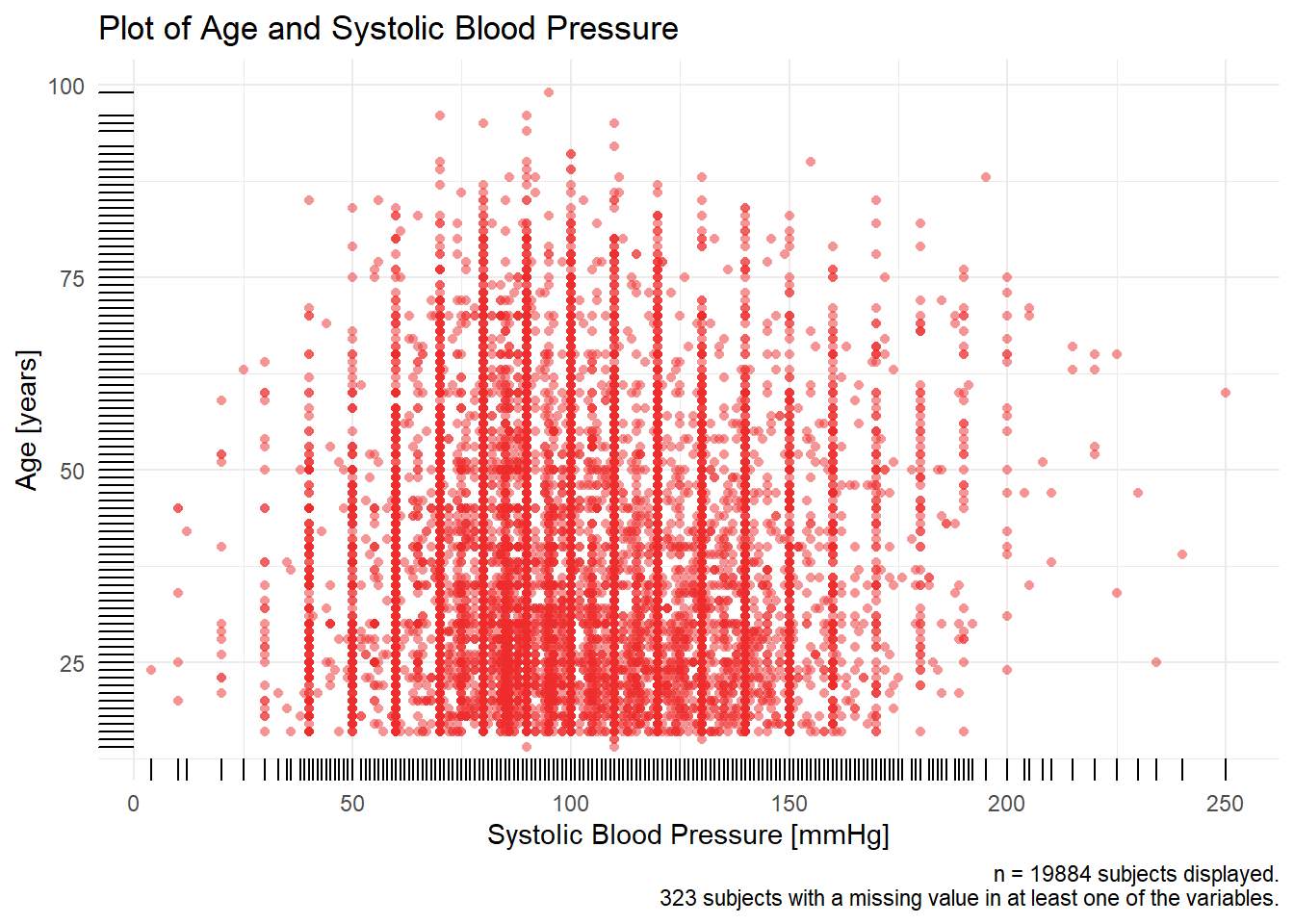
8.4 Summary reports by Glasgow coma score
| Baseline characteristics by Glasgow coma score. | ||||||||||||||
| N |
3 N=784 |
4 N=520 |
5 N=441 |
6 N=584 |
7 N=733 |
8 N=576 |
9 N=504 |
10 N=663 |
11 N=586 |
12 N=951 |
13 N=1356 |
14 N=2140 |
15 N=10346 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Age years |
20203 | 24.0 32.0 44.0 35.5 ± 14.9 |
25.0 33.0 44.0 35.5 ± 14.1 |
24.0 32.0 45.0 35.4 ± 14.7 |
23.0 31.0 45.0 35.4 ± 15.4 |
23.0 30.0 42.0 33.9 ± 14.0 |
24.0 32.0 45.0 35.7 ± 15.0 |
24.0 32.0 44.0 35.5 ± 14.6 |
24.0 31.0 42.0 34.4 ± 13.8 |
24.0 33.0 46.0 36.6 ± 15.6 |
25.0 32.0 45.0 35.9 ± 14.3 |
25.0 33.0 45.0 36.4 ± 15.0 |
24.0 31.0 44.0 35.1 ± 14.7 |
23.0 30.0 41.0 33.7 ± 13.8 |
Heart Rate /min |
20070 | 90.0 112.0 128.0 106.9 ± 31.3 |
95.0 114.0 130.0 110.8 ± 29.2 |
98.0 110.5 130.0 111.4 ± 25.4 |
90.0 110.0 123.2 106.2 ± 24.4 |
95.0 109.0 120.0 107.1 ± 23.0 |
95.0 110.0 120.0 107.4 ± 24.0 |
92.0 109.0 120.0 105.5 ± 21.9 |
96.0 110.0 124.8 108.4 ± 24.0 |
96.0 110.0 122.0 107.8 ± 20.4 |
100.0 110.0 122.0 109.3 ± 20.2 |
96.0 108.0 120.0 106.5 ± 20.1 |
92.0 105.0 120.0 104.5 ± 19.9 |
90.0 100.0 115.0 102.0 ± 18.9 |
Respiratory Rate /min |
20016 | 12.00 20.00 28.00 20.67 ± 10.74 |
16.00 22.00 28.00 22.22 ± 9.14 |
18.00 22.00 28.00 22.89 ± 8.69 |
18.00 21.00 26.00 22.12 ± 7.56 |
18.00 20.00 26.00 21.97 ± 7.69 |
18.00 22.00 28.00 23.11 ± 7.73 |
20.00 24.00 28.00 23.23 ± 6.99 |
19.00 22.00 28.00 23.05 ± 6.73 |
20.00 23.00 28.00 23.45 ± 6.37 |
20.00 24.00 28.00 24.32 ± 6.41 |
20.00 22.00 27.00 23.45 ± 6.53 |
20.00 22.00 26.00 23.41 ± 6.09 |
20.00 22.00 26.00 23.14 ± 6.07 |
Systolic Blood Pressure mmHg |
19887 | 70.0 85.0 103.0 88.7 ± 33.7 |
78.0 90.0 116.0 96.5 ± 31.2 |
80.0 90.0 118.0 99.0 ± 30.7 |
80.0 100.0 127.0 104.3 ± 32.1 |
80.0 100.0 130.0 105.4 ± 30.6 |
80.0 90.0 115.0 99.2 ± 29.4 |
80.0 96.0 120.0 99.6 ± 28.9 |
80.0 90.0 110.0 92.6 ± 28.0 |
80.0 90.0 110.0 94.4 ± 26.4 |
71.0 90.0 100.0 88.4 ± 24.7 |
80.0 90.0 110.0 95.9 ± 23.5 |
80.0 90.0 110.0 96.4 ± 22.8 |
90.0 100.0 110.0 100.5 ± 23.1 |
Central Capillary Refille Time s |
19596 | 3.00 4.00 5.00 4.15 ± 2.13 |
3.00 4.00 5.00 3.84 ± 1.90 |
2.00 3.00 5.00 3.76 ± 1.91 |
2.00 3.00 4.00 3.49 ± 1.64 |
2.00 3.00 4.00 3.28 ± 1.55 |
2.00 3.00 4.00 3.52 ± 1.69 |
2.00 3.00 4.00 3.40 ± 3.00 |
2.00 3.00 4.00 3.37 ± 1.66 |
2.00 3.00 4.00 3.27 ± 1.51 |
3.00 3.00 4.00 3.53 ± 1.60 |
2.00 3.00 4.00 3.40 ± 1.69 |
2.00 3.00 4.00 3.31 ± 1.73 |
2.00 3.00 4.00 3.06 ± 1.54 |
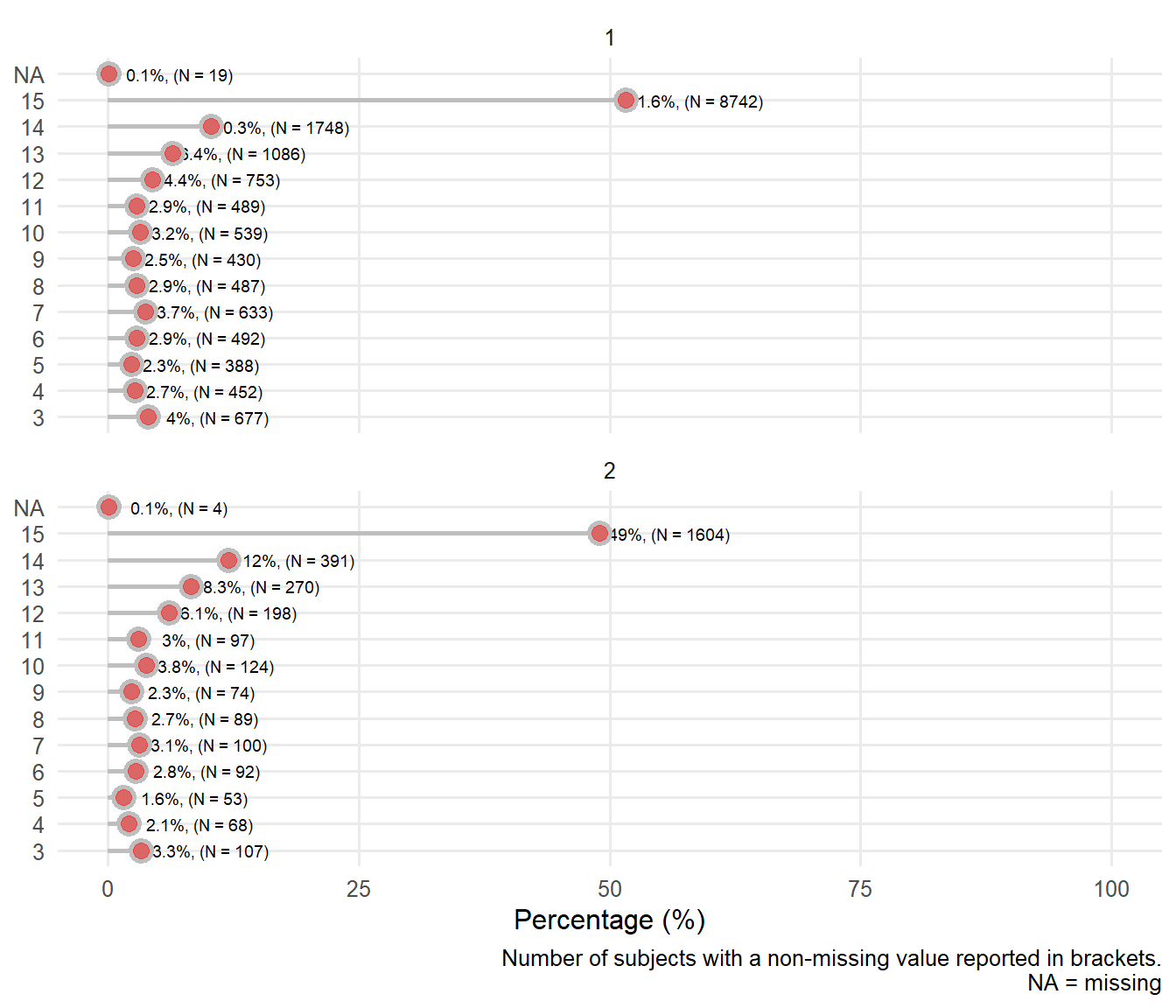
| Sex : female | 20206 | 0.14 107/ 784 | 0.13 68/ 520 | 0.12 53/ 441 | 0.16 92/ 584 | 0.14 100/ 733 | 0.15 89/ 576 | 0.15 74/ 504 | 0.19 124/ 663 | 0.17 97/ 586 | 0.21 198/ 951 | 0.20 270/ 1356 | 0.18 391/ 2139 | 0.16 1604/10346 |
Hours Since Injury hours |
20196 | 1.00 2.00 4.00 2.54 ± 1.94 |
1.00 3.00 5.00 3.26 ± 2.20 |
1.00 3.00 5.00 3.42 ± 2.19 |
2.00 3.75 6.00 3.75 ± 2.31 |
2.00 3.00 5.00 3.62 ± 2.20 |
1.00 3.00 5.00 3.30 ± 2.17 |
1.00 3.00 5.00 3.12 ± 2.20 |
1.00 2.00 4.00 3.03 ± 2.19 |
1.00 2.50 4.00 3.01 ± 2.05 |
1.00 2.00 4.00 2.75 ± 2.03 |
1.00 2.00 4.00 2.79 ± 1.97 |
1.00 2.00 4.00 2.64 ± 1.99 |
1.00 2.00 4.00 2.71 ± 2.69 |
| Injury type : blunt | 20207 | 0.62 483/ 784 | 0.71 371/ 520 | 0.73 324/ 441 | 0.76 443/ 584 | 0.76 559/ 733 | 0.69 399/ 576 | 0.67 338/ 504 | 0.61 407/ 663 | 0.64 377/ 586 | 0.58 550/ 951 | 0.60 814/ 1356 | 0.58 1237/ 2140 | 0.47 4880/10346 |
| penetrating | 0.22 175/ 784 | 0.10 53/ 520 | 0.09 41/ 441 | 0.10 59/ 584 | 0.11 77/ 733 | 0.15 89/ 576 | 0.17 88/ 504 | 0.23 151/ 663 | 0.21 123/ 586 | 0.29 272/ 951 | 0.24 326/ 1356 | 0.29 629/ 2140 | 0.43 4458/10346 | |
| blunt and penetrating | 0.16 126/ 784 | 0.18 96/ 520 | 0.17 76/ 441 | 0.14 82/ 584 | 0.13 97/ 733 | 0.15 88/ 576 | 0.15 78/ 504 | 0.16 105/ 663 | 0.15 86/ 586 | 0.14 129/ 951 | 0.16 216/ 1356 | 0.13 274/ 2140 | 0.10 1008/10346 | |
| a b c represent the lower quartile a, the median b, and the upper quartile c for continuous variables. x ± s represents X ± 1 SD. N is the number of non-missing values. | ||||||||||||||
8.4.2 Distribution of systolic blood pressure by Glasgow coma score
Figure 8.11: Distribution of systolic blood pressure by gcs
8.4.3 Distribution of heart rate by Glasgow coma score
Figure 8.12: Distribution of heart rate by gcs
8.5 Section session info
## R version 4.1.3 (2022-03-10)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_Austria.1252 LC_CTYPE=English_Austria.1252
## [3] LC_MONETARY=English_Austria.1252 LC_NUMERIC=C
## [5] LC_TIME=English_Austria.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] patchwork_1.1.1 corrplot_0.92 gtsummary_1.5.2 Hmisc_4.6-0
## [5] Formula_1.2-4 survival_3.2-13 lattice_0.20-45 plotly_4.10.0
## [9] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.8 purrr_0.3.4
## [13] readr_2.1.2 tidyr_1.2.0 tibble_3.1.6 ggplot2_3.3.5
## [17] tidyverse_1.3.1 here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] fs_1.5.2 lubridate_1.8.0 RColorBrewer_1.1-2
## [4] httr_1.4.2 rprojroot_2.0.2 tools_4.1.3
## [7] backports_1.4.1 bslib_0.3.1 utf8_1.2.2
## [10] R6_2.5.1 rpart_4.1.16 DBI_1.1.2
## [13] lazyeval_0.2.2 colorspace_2.0-3 nnet_7.3-17
## [16] withr_2.5.0 tidyselect_1.1.2 gridExtra_2.3
## [19] compiler_4.1.3 cli_3.2.0 rvest_1.0.2
## [22] gt_0.4.0 htmlTable_2.4.0 xml2_1.3.3
## [25] labeling_0.4.2 bookdown_0.25 sass_0.4.1
## [28] checkmate_2.0.0 scales_1.1.1 commonmark_1.8.0
## [31] digest_0.6.29 foreign_0.8-82 rmarkdown_2.13
## [34] base64enc_0.1-3 jpeg_0.1-9 pkgconfig_2.0.3
## [37] htmltools_0.5.2 highr_0.9 dbplyr_2.1.1
## [40] fastmap_1.1.0 htmlwidgets_1.5.4 rlang_1.0.2
## [43] readxl_1.3.1 rstudioapi_0.13 farver_2.1.0
## [46] jquerylib_0.1.4 generics_0.1.2 jsonlite_1.8.0
## [49] crosstalk_1.2.0 magrittr_2.0.2 Matrix_1.4-0
## [52] Rcpp_1.0.8.3 munsell_0.5.0 fansi_1.0.3
## [55] lifecycle_1.0.1 stringi_1.7.6 yaml_2.3.5
## [58] grid_4.1.3 crayon_1.5.1 haven_2.4.3
## [61] splines_4.1.3 hms_1.1.1 knitr_1.38
## [64] pillar_1.7.0 reprex_2.0.1 glue_1.6.2
## [67] evaluate_0.15 latticeExtra_0.6-29 broom.helpers_1.6.0
## [70] data.table_1.14.2 modelr_0.1.8 vctrs_0.3.8
## [73] png_0.1-7 tzdb_0.2.0 cellranger_1.1.0
## [76] gtable_0.3.0 assertthat_0.2.1 xfun_0.30
## [79] broom_0.7.12 viridisLite_0.4.0 cluster_2.1.2
## [82] ellipsis_0.3.2