Chapter 17 Univariate distribution checks
This section reports a series of univariate summary checks of the bacteremia dataset.
17.1 Data set overview
Using the Hmisc describe function, we provide an overview of the data set. The descriptive report also provides histograms of continuous variables. For ease of scanning the information, we group the report by measurement type.
17.1.1 Demographic variables
2 Variables 14691 Observations
Alter: Patient Age years
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14691 | 0 | 85 | 1 | 56.17 | 20.78 | 24 | 29 | 43 | 58 | 70 | 79 | 84 |
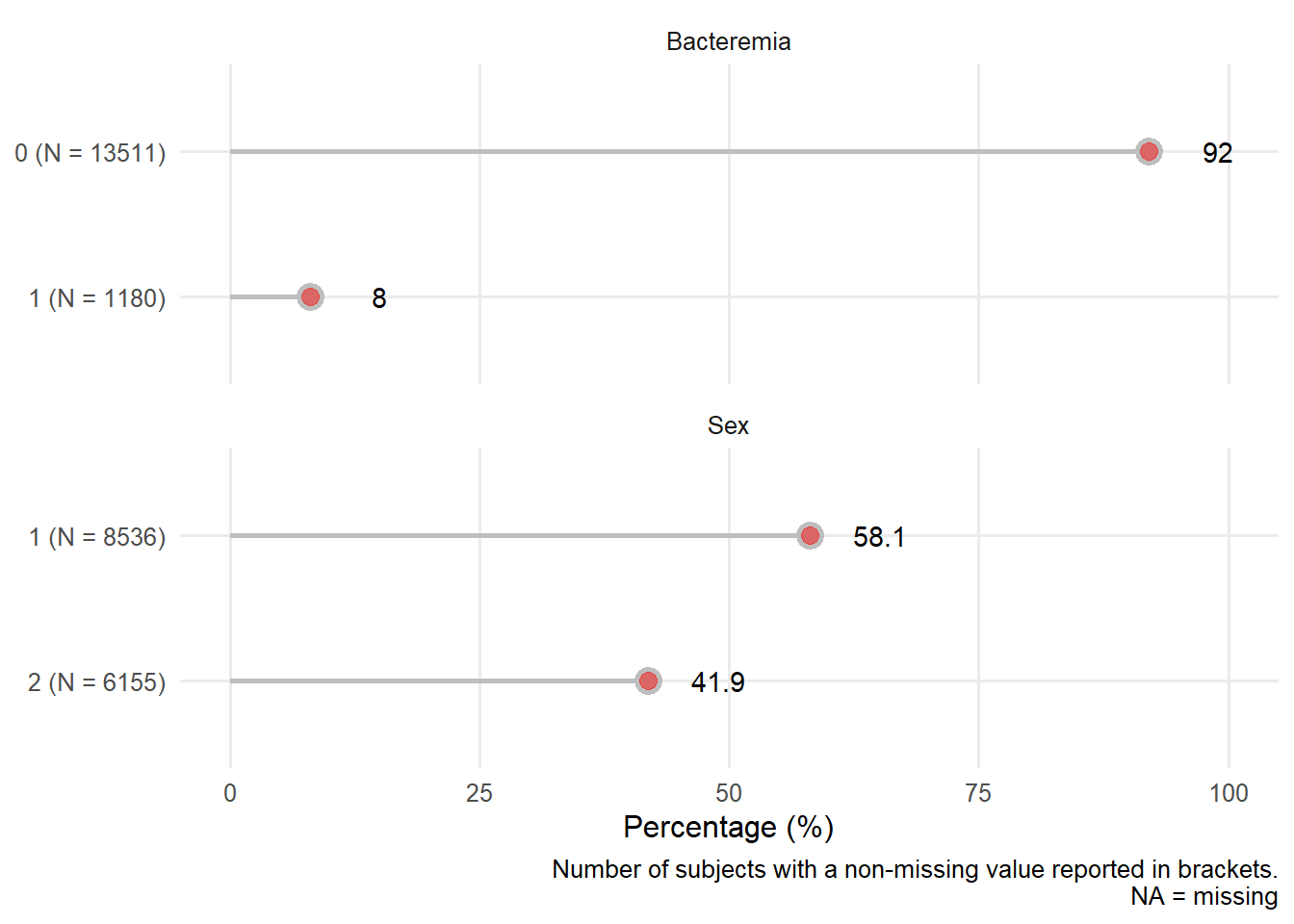
sex: Patient Sex 1=male, 2=female
| n | missing | distinct | Info | Mean | Gmd |
|---|---|---|---|---|---|
| 14691 | 0 | 2 | 0.73 | 1.419 | 0.4869 |
Value 1 2 Frequency 8536 6155 Proportion 0.581 0.419
17.1.2 Pivotal variables and very important predictors
6 Variables 14691 Observations
WBC: White blood count G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14229 | 462 | 2710 | 1 | 11.23 | 7.602 | 2.66 | 4.26 | 6.63 | 9.60 | 13.53 | 18.22 | 22.27 |
Alter: Patient Age years
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14691 | 0 | 85 | 1 | 56.17 | 20.78 | 24 | 29 | 43 | 58 | 70 | 79 | 84 |
BUN: Blood urea nitrogen mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14519 | 172 | 947 | 1 | 22.66 | 16.92 | 7.1 | 8.6 | 11.6 | 16.6 | 26.9 | 44.8 | 60.8 |
KREA: Creatinine mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14532 | 159 | 674 | 1 | 1.329 | 0.8518 | 0.620 | 0.690 | 0.810 | 1.000 | 1.350 | 2.160 | 3.144 |
NEU: Neutrophiles G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13963 | 728 | 374 | 1 | 8.367 | 5.776 | 1.60 | 2.70 | 4.60 | 7.30 | 10.80 | 15.08 | 18.40 |
PLT: Blood platelets G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14649 | 42 | 718 | 1 | 220 | 130.1 | 50 | 81 | 140 | 204 | 277 | 369 | 445 |
17.1.6 Remaining variables
29 Variables 14691 Observations
MCV: Mean corpuscular volume pg
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14649 | 42 | 506 | 1 | 88.35 | 6.992 | 78.2 | 81.1 | 84.7 | 88.3 | 92.0 | 95.9 | 99.0 |
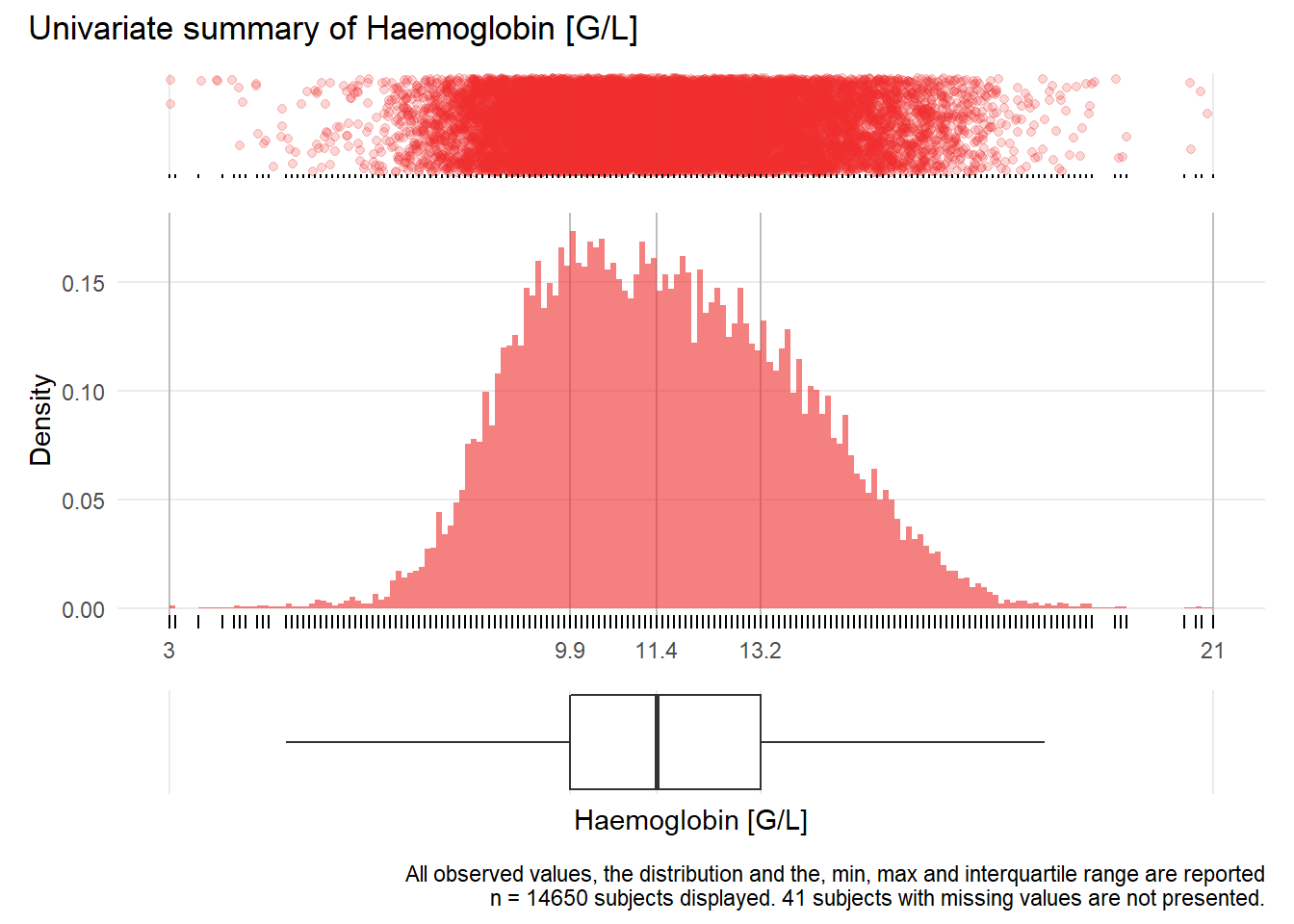
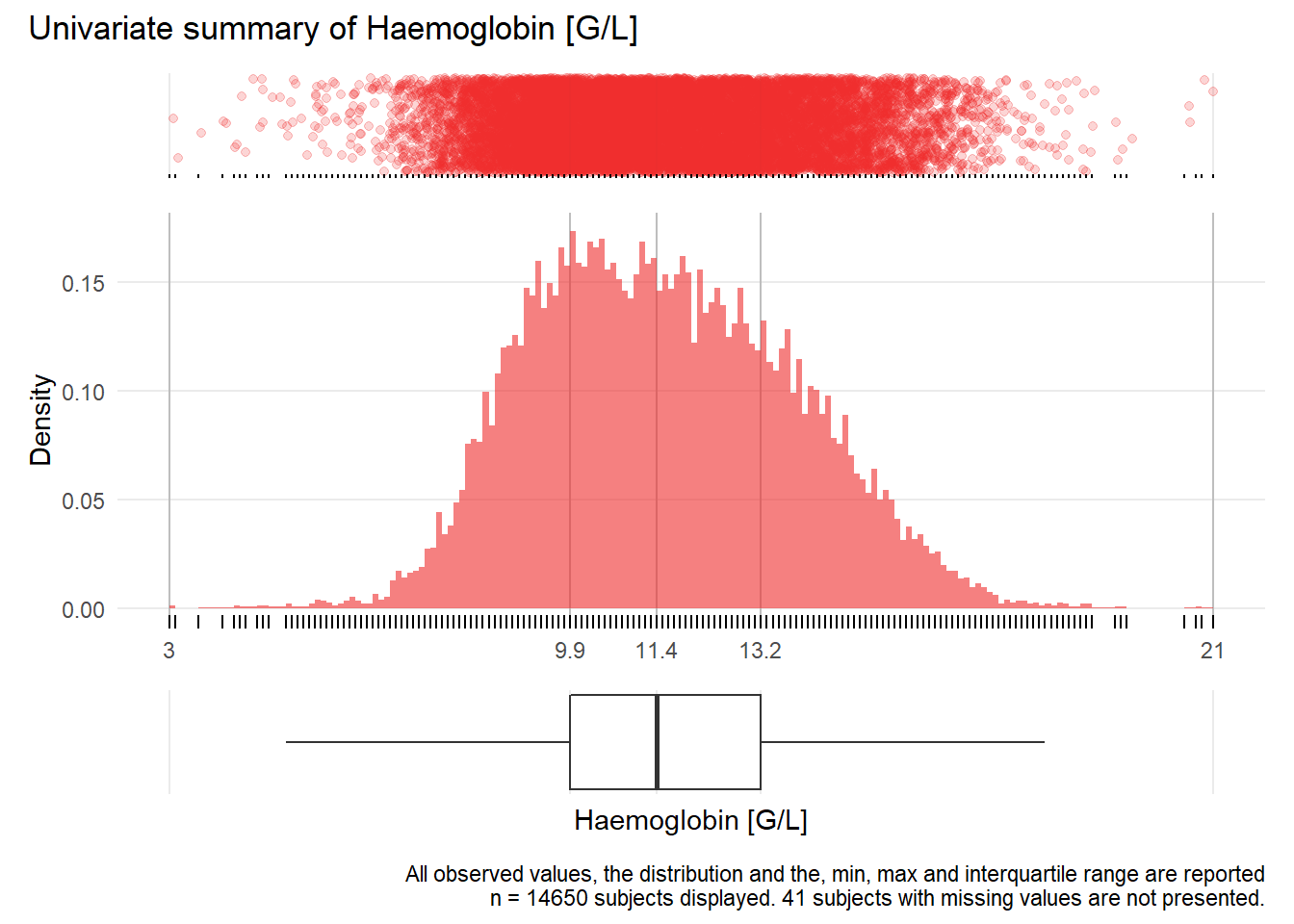
HGB: Haemoglobin G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14650 | 41 | 157 | 1 | 11.57 | 2.558 | 8.2 | 8.8 | 9.9 | 11.4 | 13.2 | 14.6 | 15.4 |
HCT: Haematocrit %
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14649 | 42 | 404 | 1 | 34.48 | 7.316 | 24.6 | 26.4 | 29.8 | 34.3 | 39.1 | 42.9 | 44.8 |
MCH: Mean corpuscular hemoglobin fl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14649 | 42 | 232 | 1 | 29.58 | 2.693 | 25.3 | 26.7 | 28.4 | 29.7 | 31.0 | 32.4 | 33.4 |
MCHC: Mean corpuscular hemoglobin concentration g/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14649 | 42 | 124 | 0.999 | 33.47 | 1.546 | 31.1 | 31.7 | 32.6 | 33.5 | 34.4 | 35.2 | 35.6 |
RDW: Red blood cell distribution width %
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14635 | 56 | 173 | 1 | 15 | 2.385 | 12.4 | 12.7 | 13.4 | 14.5 | 16.0 | 18.0 | 19.5 |
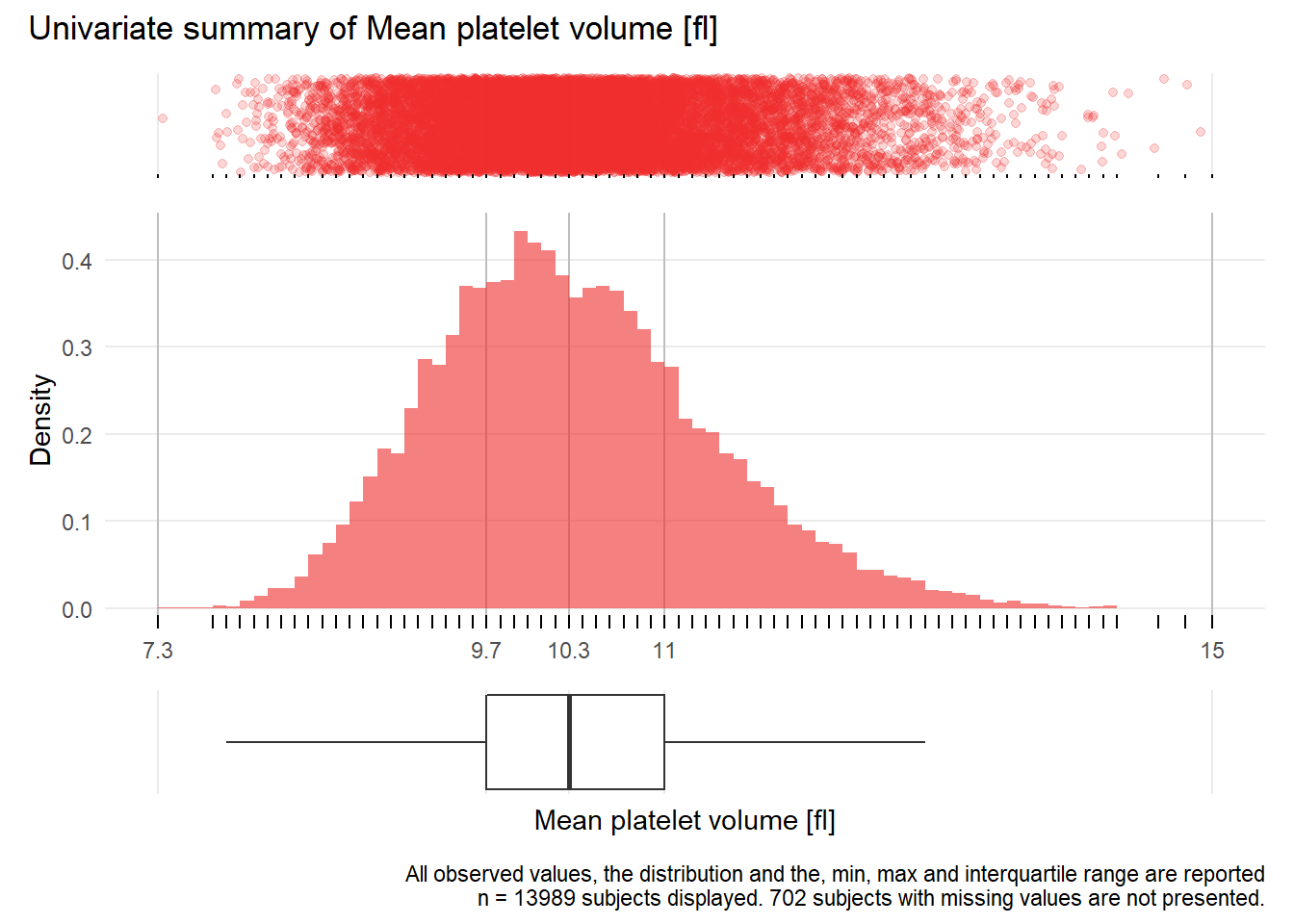
MPV: Mean platelet volume fl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13989 | 702 | 71 | 0.999 | 10.38 | 1.132 | 8.9 | 9.2 | 9.7 | 10.3 | 11.0 | 11.7 | 12.2 |
NT: Normotest %
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12224 | 2467 | 149 | 1 | 83.22 | 30.56 | 35 | 48 | 67 | 83 | 101 | 118 | 128 |
APTT: Activated partial thromboplastin time sec
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12142 | 2549 | 631 | 1 | 40.06 | 9.533 | 30.1 | 31.4 | 34.1 | 37.7 | 42.7 | 49.9 | 56.6 |
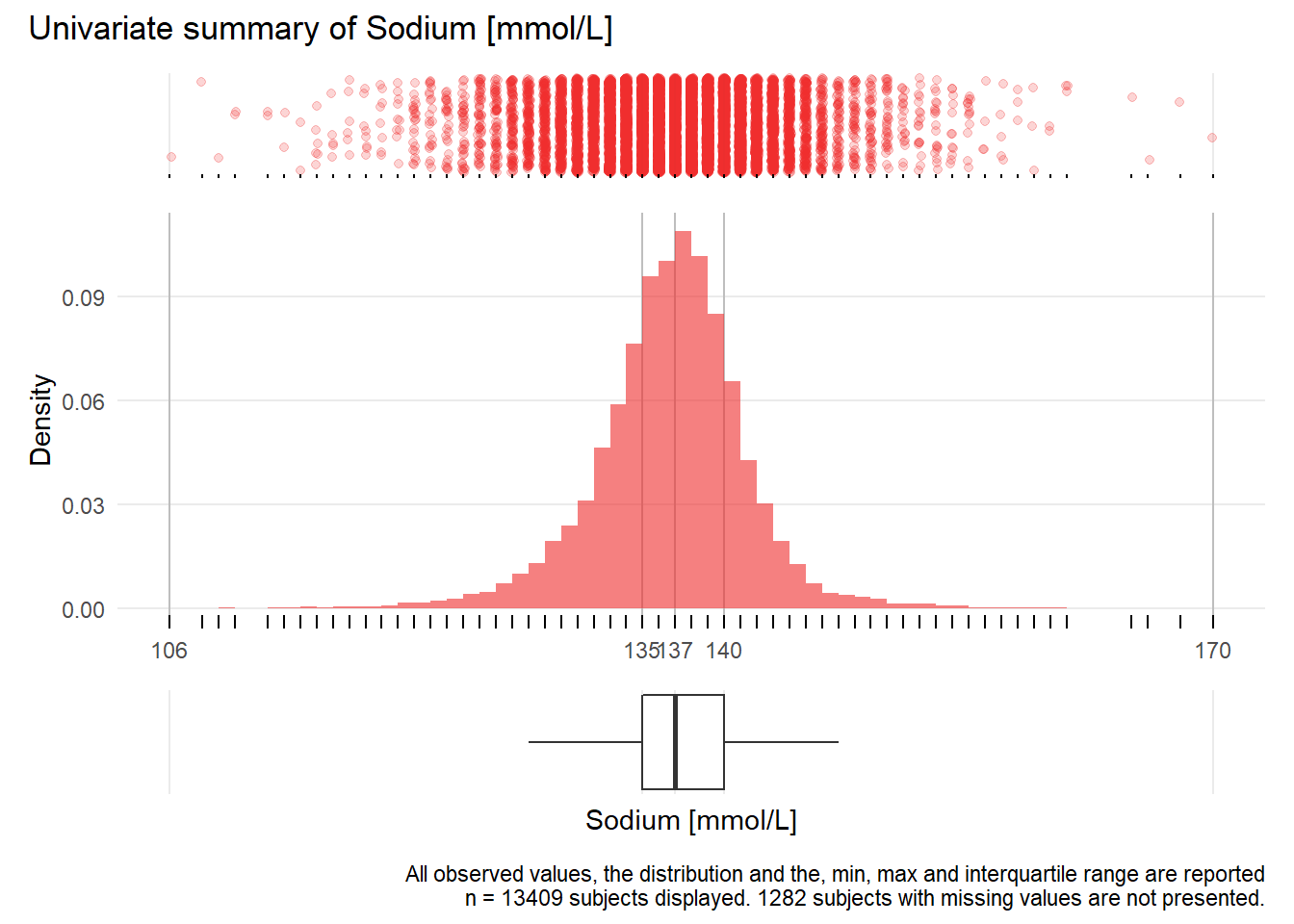
NA.: Sodium mmol/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13409 | 1282 | 58 | 0.994 | 137.2 | 5.034 | 129 | 132 | 135 | 137 | 140 | 142 | 144 |
CA: Calcium mmol/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13415 | 1276 | 185 | 1 | 2.214 | 0.2213 | 1.89 | 1.96 | 2.09 | 2.22 | 2.35 | 2.45 | 2.51 |
PHOS: Phosphate mmol/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13449 | 1242 | 306 | 1 | 1.048 | 0.3993 | 0.55 | 0.64 | 0.81 | 0.99 | 1.20 | 1.47 | 1.74 |
MG: Magnesium mmol/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12822 | 1869 | 146 | 0.999 | 0.8136 | 0.1609 | 0.59 | 0.64 | 0.72 | 0.81 | 0.89 | 0.98 | 1.06 |
HS: Uric acid mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11630 | 3061 | 169 | 1 | 5.413 | 2.625 | 2.2 | 2.7 | 3.7 | 5.0 | 6.6 | 8.5 | 10.0 |
GBIL: Bilirubin mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13250 | 1441 | 885 | 1 | 1.406 | 1.477 | 0.33 | 0.39 | 0.53 | 0.77 | 1.23 | 2.34 | 3.96 |
TP: Total protein G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13108 | 1583 | 649 | 1 | 64.9 | 12.97 | 45.20 | 49.47 | 56.90 | 65.70 | 73.30 | 78.80 | 82.00 |
ALB: Albumin G/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13015 | 1676 | 401 | 1 | 33.42 | 8.513 | 21.3 | 23.6 | 27.9 | 33.6 | 39.1 | 43.2 | 45.2 |
AMY: Amylase U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10778 | 3913 | 488 | 1 | 90.83 | 100.5 | 18 | 23 | 33 | 49 | 76 | 125 | 187 |
Value 0 500 1000 1500 2000 2500 4000 4500 5000 40500 44000 56000 Frequency 10432 268 39 14 12 4 2 2 2 1 1 1 Proportion 0.968 0.025 0.004 0.001 0.001 0.000 0.000 0.000 0.000 0.000 0.000 0.000For the frequency table, variable is rounded to the nearest 500
PAMY: Pancreas amylase U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7577 | 7114 | 280 | 0.999 | 41.66 | 47.28 | 7 | 9 | 14 | 22 | 36 | 64 | 97 |
Value 0 500 1000 1500 2000 3000 38500 Frequency 7495 65 7 6 2 1 1 Proportion 0.989 0.009 0.001 0.001 0.000 0.000 0.000For the frequency table, variable is rounded to the nearest 500
LIP: Lipases U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10992 | 3699 | 444 | 1 | 63.82 | 89.88 | 6 | 8 | 14 | 23 | 40 | 79 | 135 |
CHE: Cholinesterase kU/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12244 | 2447 | 997 | 1 | 4.79 | 2.378 | 1.70 | 2.17 | 3.15 | 4.60 | 6.22 | 7.65 | 8.49 |
AP: Alkaline phosphatase U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13291 | 1400 | 672 | 1 | 118.8 | 91.51 | 42 | 49 | 63 | 84 | 123 | 206 | 302 |
LDH: Lactate dehydrogenase U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12977 | 1714 | 1137 | 1 | 331.2 | 240.9 | 136 | 152 | 187 | 239 | 332 | 508 | 724 |
CK: Creatinine kinases U/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12611 | 2080 | 1506 | 1 | 385 | 615.4 | 18 | 25 | 42 | 80 | 184 | 577 | 1155 |
GLU: Glucoses mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10499 | 4192 | 389 | 1 | 126.4 | 48.3 | 78 | 85 | 97 | 113 | 138 | 177 | 216 |
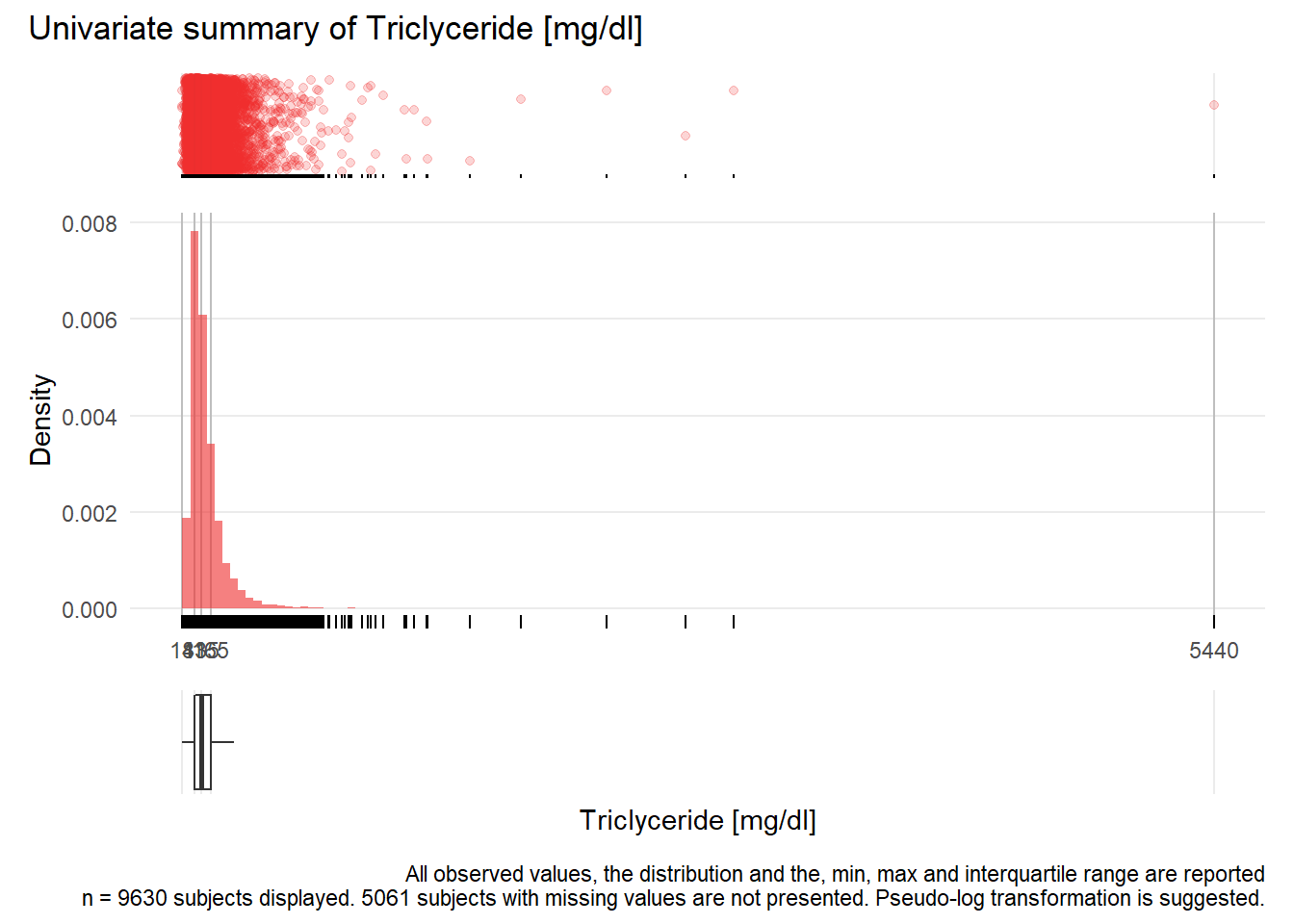
TRIG: Triclyceride mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9630 | 5061 | 538 | 1 | 141.7 | 90.33 | 54 | 64 | 83 | 115 | 165 | 241 | 307 |
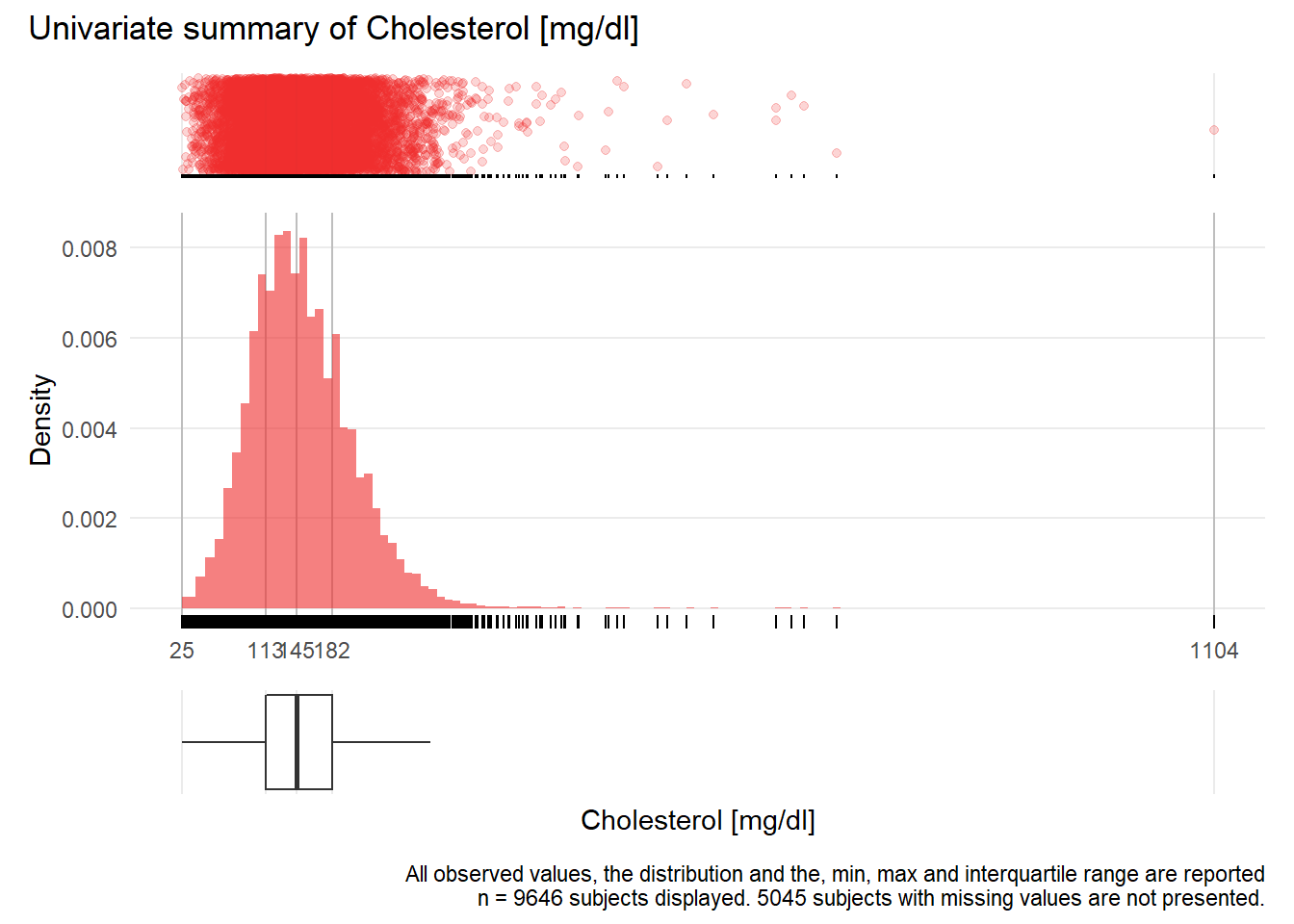
CHOL: Cholesterol mg/dl
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9646 | 5045 | 339 | 1 | 150.8 | 59.23 | 74 | 89 | 113 | 145 | 182 | 219 | 243 |
PDW: Platelet distribution width %
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13589 | 1102 | 167 | 1 | 12.29 | 2.375 | 9.3 | 9.8 | 10.8 | 12.0 | 13.4 | 15.1 | 16.4 |
RBC: Red blood count T/L
| n | missing | distinct | Info | Mean | Gmd | .05 | .10 | .25 | .50 | .75 | .90 | .95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14230 | 461 | 65 | 0.999 | 3.936 | 0.8772 | 2.7 | 2.9 | 3.4 | 3.9 | 4.5 | 4.9 | 5.2 |
17.2 Categorical variables
We now provide a closer visual examination of the categorical predictors.
17.3 Continuous variables
17.3.1 Suggested transformations
Next we investigate whether a transformation of continuous variables may improve any further analyses to reduce disproportional impact of highly influential points, also in multivariate summaries. We employ a function ida_trans for this purpose, which optimises the parameter sigma of the pseudo-logarithm for that purpose. The optimization targets the best possible linear correlation of the transformed values with normal deviates. If no better transformation can be found, no transformation is suggested.
variables<- c("Alter", pivotal_vars, vip_vars, leuko_related_vars, leuko_ratio_vars, kidney_related_vars, acute_related_vars, remaining_vars)
unique.variables <- unique(variables)
#variables<- c("Alter", pivotal_vars, vip_vars)
res<-sapply(unique.variables, function(X) ida_trans(b_bact[,X])$const) #takes long, calculate once, and save?
res## Alter WBC BUN KREA NEU PLT EOS
## NA 2.51471408 0.03198339 0.03193846 2.30783002 NA 0.11873740
## BASO LYM MONO NEUR EOSR BASOR LYMR
## 0.12980073 0.17957981 0.26505156 NA 0.42874860 0.17300902 1.77333947
## MONOR K eGFR BUN_KREA FIB CRP ASAT
## 3.00040692 0.02677349 NA 0.03194543 NA NA 0.03185536
## ALAT GGT MCV HGB HCT MCH MCHC
## 1.02570312 0.03185702 NA NA NA NA NA
## RDW MPV NT APTT NA. CA PHOS
## NA NA NA 0.03047767 NA NA 0.14534462
## MG HS GBIL TP ALB AMY PAMY
## NA NA 0.03306450 NA NA 0.02970893 0.03005131
## LIP CHE AP LDH CK GLU TRIG
## 1.02558160 NA 0.02888640 0.02191602 0.02786388 0.01875994 0.02911146
## CHOL PDW RBC
## NA NA NARegister transformed variables in the data set:
for(j in 1:length(unique.variables)){
if(!is.na(res[j])){
newname <- paste("t_",unique.variables[j],sep="")
newlabel <- paste("pseudo-log of",label(b_bact)[unique.variables[j]])
names(newlabel)<-newname
x<-pseudo_log(b_bact[[unique.variables[j]]], sigma=res[j], base=10)
label(x)<-newlabel
b_bact[[newname]] <- x
upData(b_bact, labels=newlabel)
}
}## Input object size: 5575040 bytes; 57 variables 14691 observations
## New object size: 5574816 bytes; 57 variables 14691 observations
## Input object size: 5693696 bytes; 58 variables 14691 observations
## New object size: 5693472 bytes; 58 variables 14691 observations
## Input object size: 5812336 bytes; 59 variables 14691 observations
## New object size: 5812112 bytes; 59 variables 14691 observations
## Input object size: 5930976 bytes; 60 variables 14691 observations
## New object size: 5930752 bytes; 60 variables 14691 observations
## Input object size: 6049616 bytes; 61 variables 14691 observations
## New object size: 6049392 bytes; 61 variables 14691 observations
## Input object size: 6168256 bytes; 62 variables 14691 observations
## New object size: 6168032 bytes; 62 variables 14691 observations
## Input object size: 6286896 bytes; 63 variables 14691 observations
## New object size: 6286672 bytes; 63 variables 14691 observations
## Input object size: 6405536 bytes; 64 variables 14691 observations
## New object size: 6405312 bytes; 64 variables 14691 observations
## Input object size: 6524176 bytes; 65 variables 14691 observations
## New object size: 6523952 bytes; 65 variables 14691 observations
## Input object size: 6642816 bytes; 66 variables 14691 observations
## New object size: 6642592 bytes; 66 variables 14691 observations
## Input object size: 6761464 bytes; 67 variables 14691 observations
## New object size: 6761240 bytes; 67 variables 14691 observations
## Input object size: 6880104 bytes; 68 variables 14691 observations
## New object size: 6879880 bytes; 68 variables 14691 observations
## Input object size: 6998744 bytes; 69 variables 14691 observations
## New object size: 6998520 bytes; 69 variables 14691 observations
## Input object size: 7117408 bytes; 70 variables 14691 observations
## New object size: 7117176 bytes; 70 variables 14691 observations
## Input object size: 7236064 bytes; 71 variables 14691 observations
## New object size: 7235840 bytes; 71 variables 14691 observations
## Input object size: 7354720 bytes; 72 variables 14691 observations
## New object size: 7354496 bytes; 72 variables 14691 observations
## Input object size: 7473376 bytes; 73 variables 14691 observations
## New object size: 7473152 bytes; 73 variables 14691 observations
## Input object size: 7592048 bytes; 74 variables 14691 observations
## New object size: 7591824 bytes; 74 variables 14691 observations
## Input object size: 7710688 bytes; 75 variables 14691 observations
## New object size: 7710464 bytes; 75 variables 14691 observations
## Input object size: 7829328 bytes; 76 variables 14691 observations
## New object size: 7829104 bytes; 76 variables 14691 observations
## Input object size: 7947968 bytes; 77 variables 14691 observations
## New object size: 7947744 bytes; 77 variables 14691 observations
## Input object size: 8066608 bytes; 78 variables 14691 observations
## New object size: 8066384 bytes; 78 variables 14691 observations
## Input object size: 8185248 bytes; 79 variables 14691 observations
## New object size: 8185024 bytes; 79 variables 14691 observations
## Input object size: 8303904 bytes; 80 variables 14691 observations
## New object size: 8303680 bytes; 80 variables 14691 observations
## Input object size: 8422560 bytes; 81 variables 14691 observations
## New object size: 8422336 bytes; 81 variables 14691 observations
## Input object size: 8541216 bytes; 82 variables 14691 observations
## New object size: 8540992 bytes; 82 variables 14691 observations
## Input object size: 8659856 bytes; 83 variables 14691 observations
## New object size: 8659632 bytes; 83 variables 14691 observations
## Input object size: 8778496 bytes; 84 variables 14691 observations
## New object size: 8778272 bytes; 84 variables 14691 observationssigma_values <- res
c_bact <- b_bact
# update variable lists - generate a second list with transformed variables replacing the originals
bact_transformed <- bact_variables
for(j in 1:length(bact_variables)){
for(jj in 1:length(bact_variables[[j]])){
if(!is.na(res[bact_variables[[j]][jj]])) bact_transformed[[j]][jj] <- paste("t_", bact_variables[[j]][jj], sep="")
}
}17.3.2 Univariate distribution with variables using the original variable and the suggested transformations
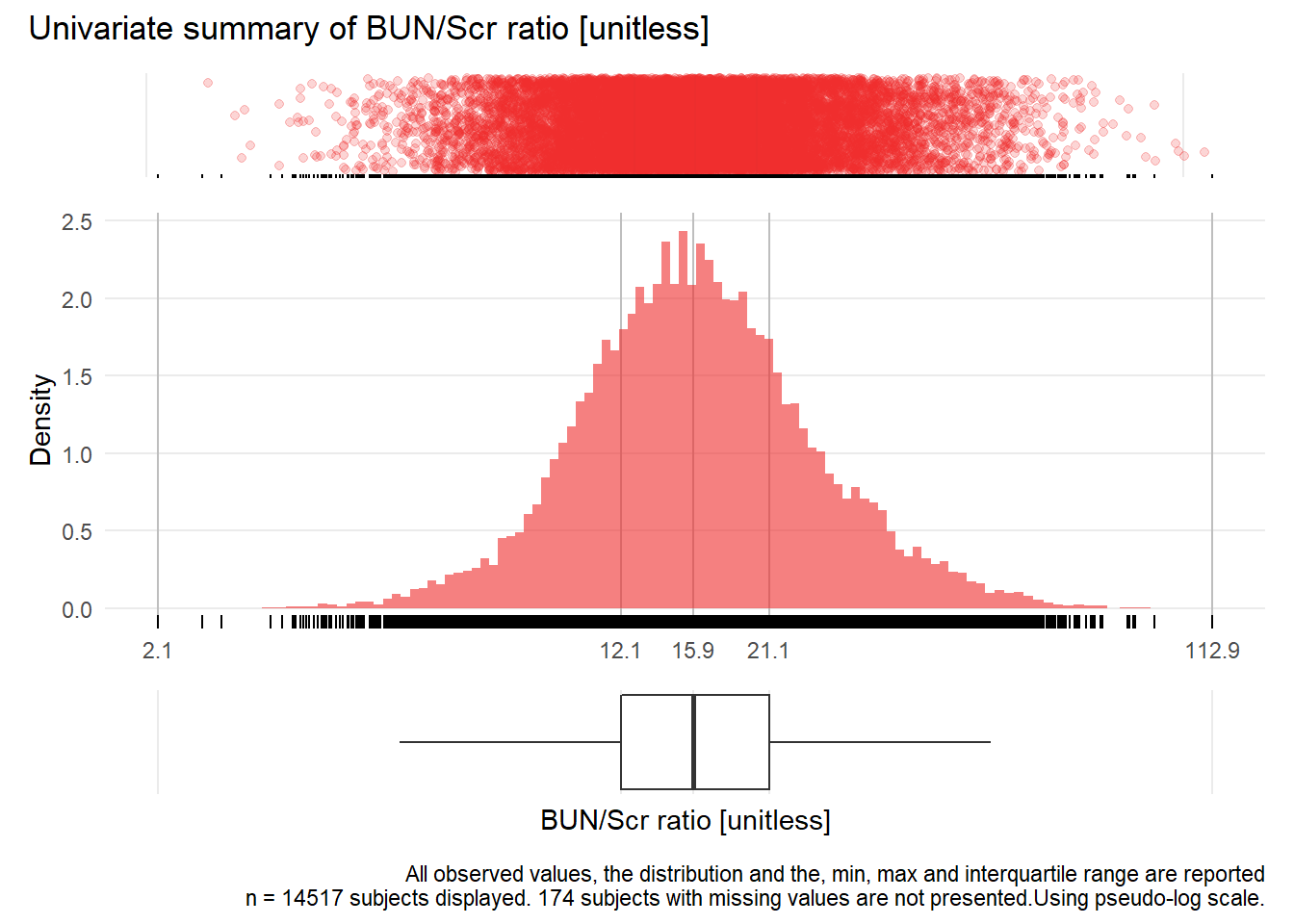
for(j in 1:length(unique.variables)){
print(ida_plot_univar(b_bact, unique.variables[j], sigma=res[j], n_bars=100))
# if(!is.na(res[j])){
# print(ida_plot_univar(b_bact, paste("t_",variables[j],sep="")))
# }
}## Warning: Removed 4 rows containing missing values (geom_point).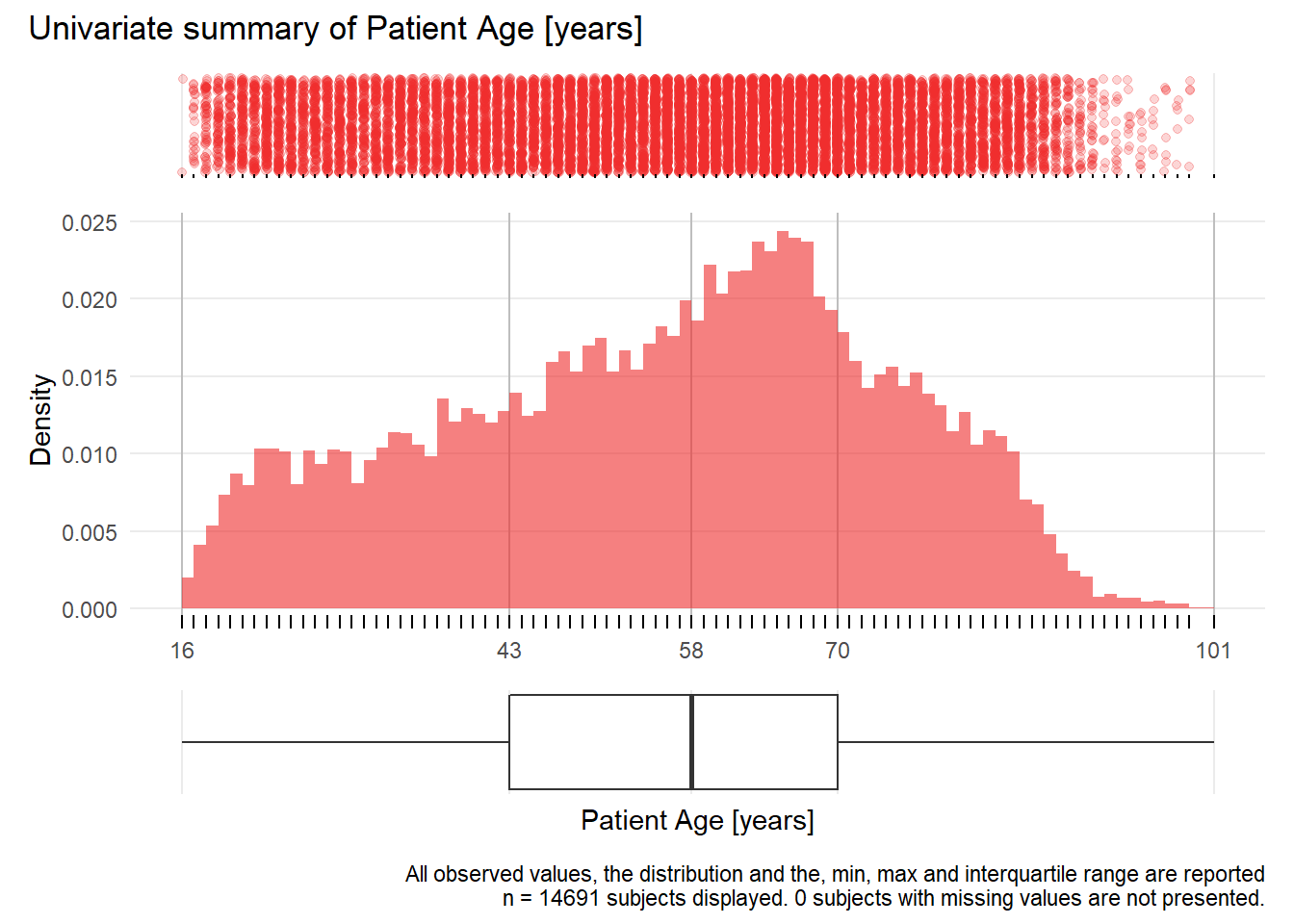
## Warning: Removed 95 rows containing missing values (geom_point).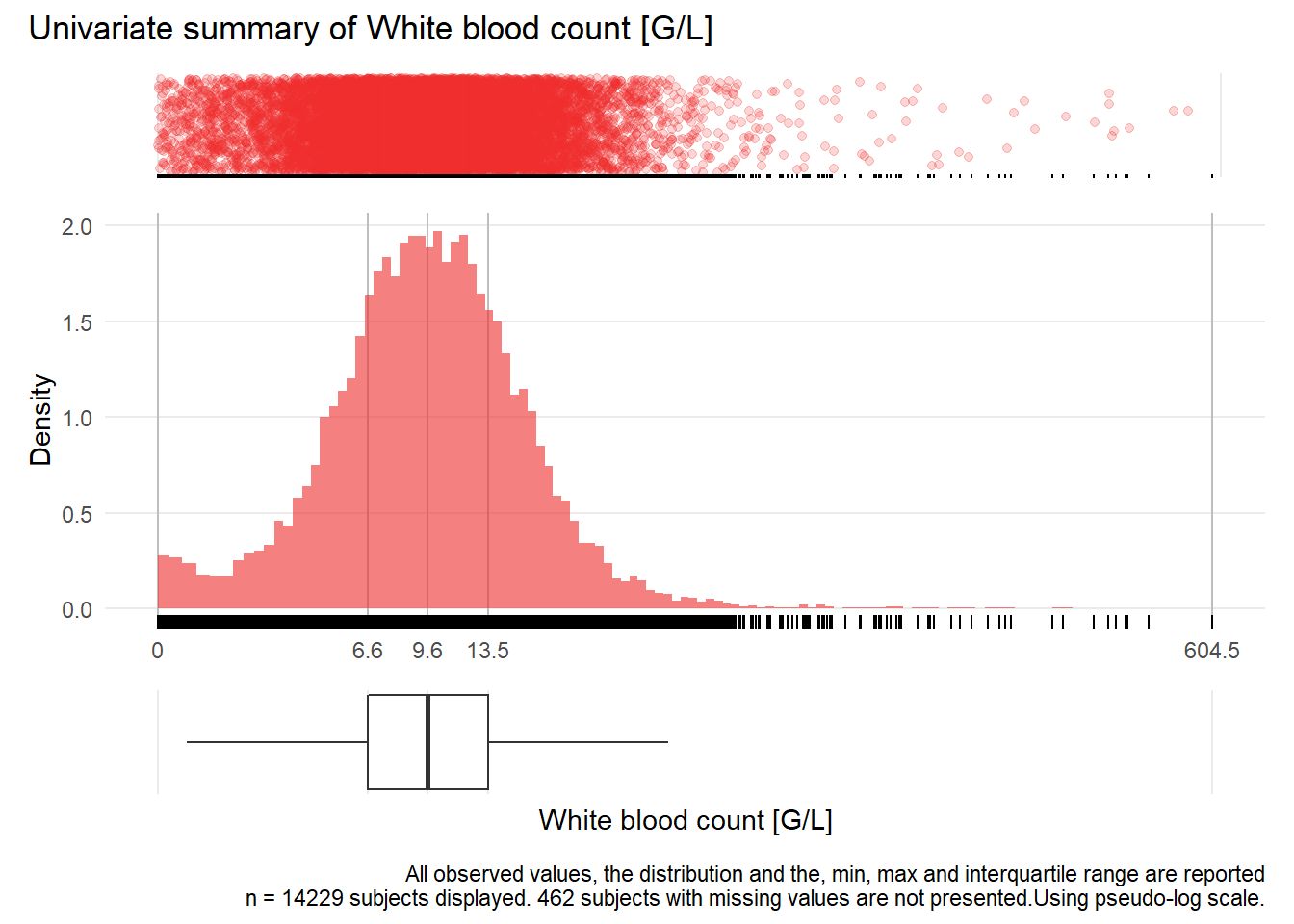
## Warning: Removed 3 rows containing missing values (geom_point).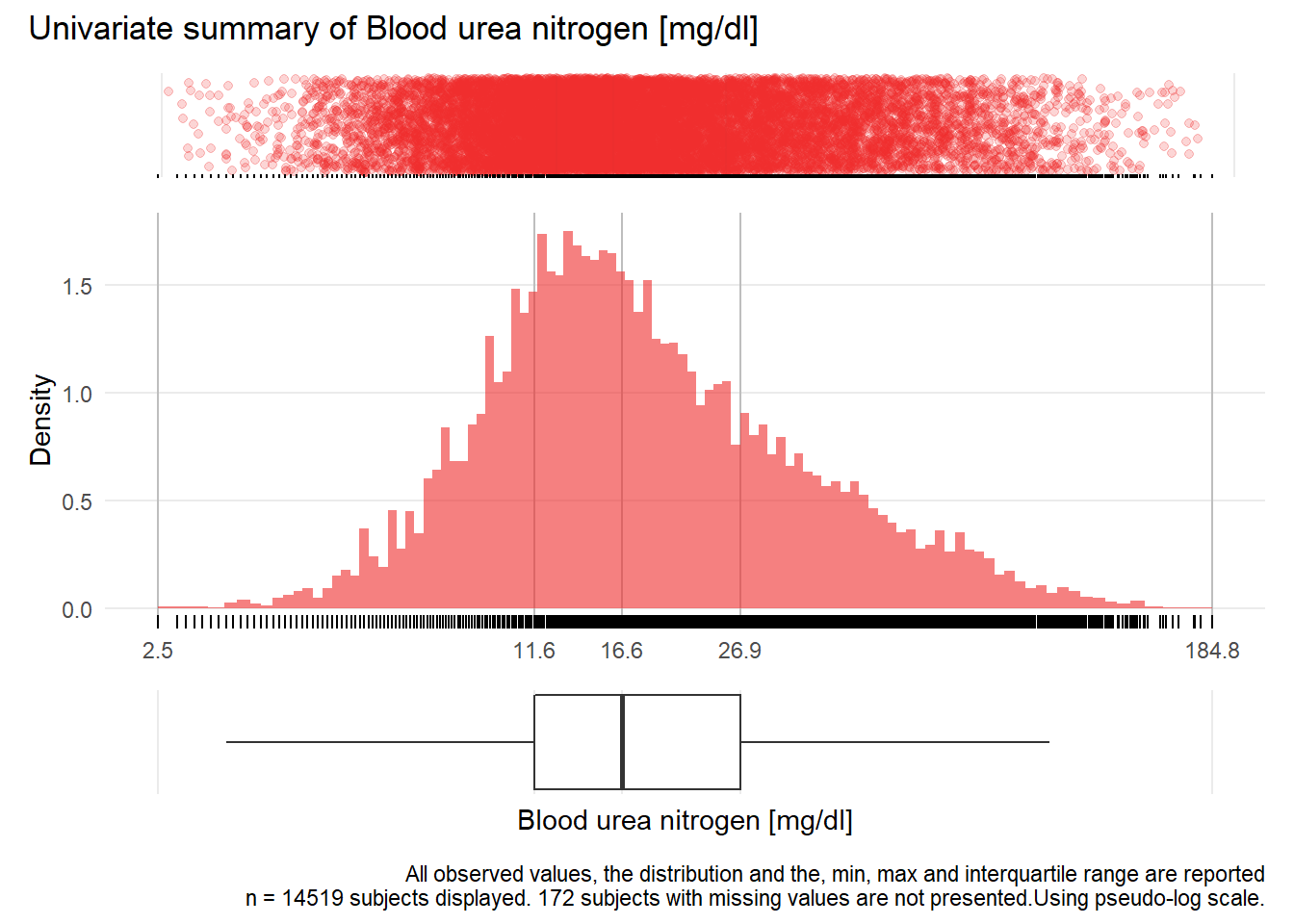
## Warning: Removed 5 rows containing missing values (geom_point).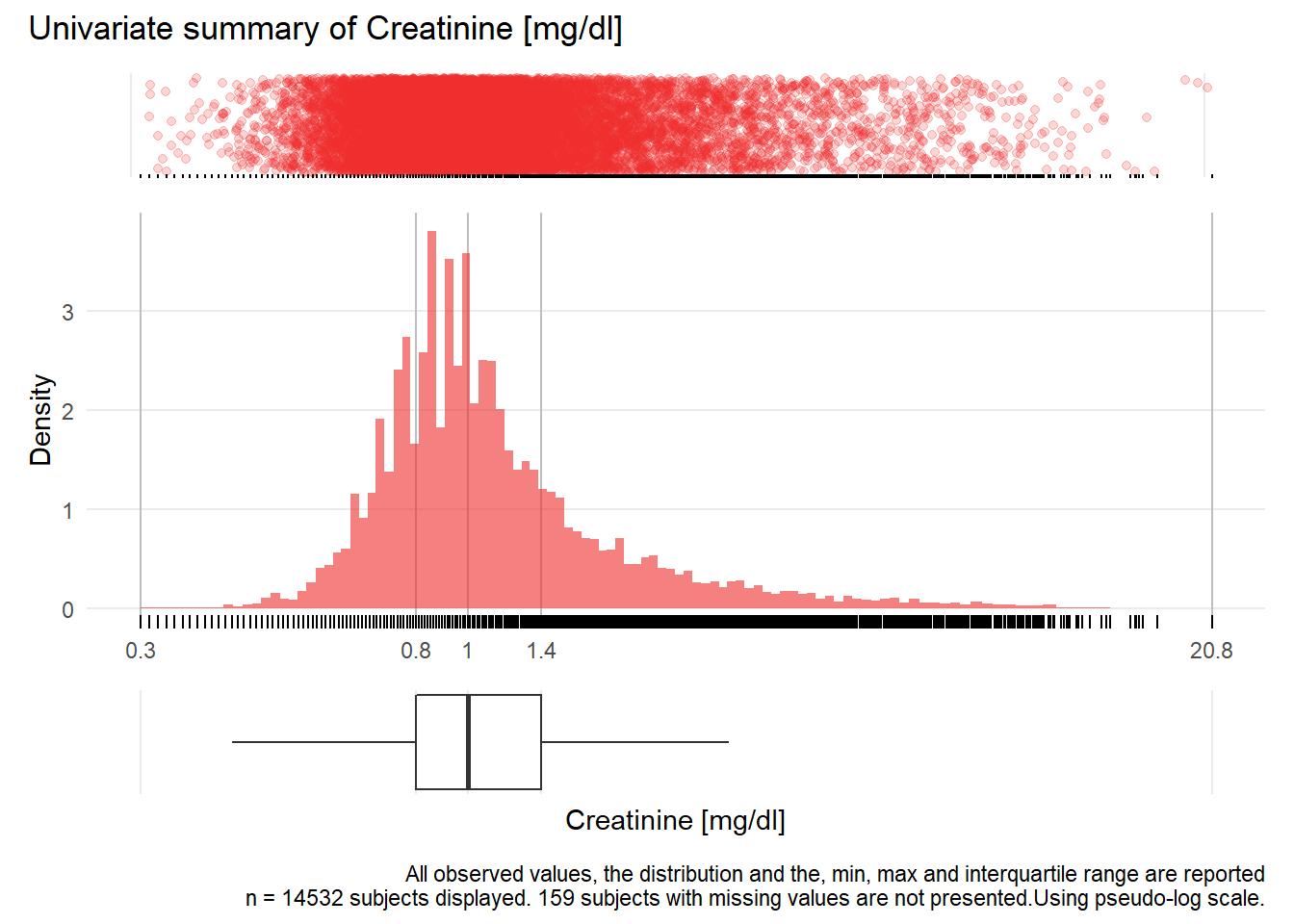
## Warning: Removed 162 rows containing missing values (geom_point).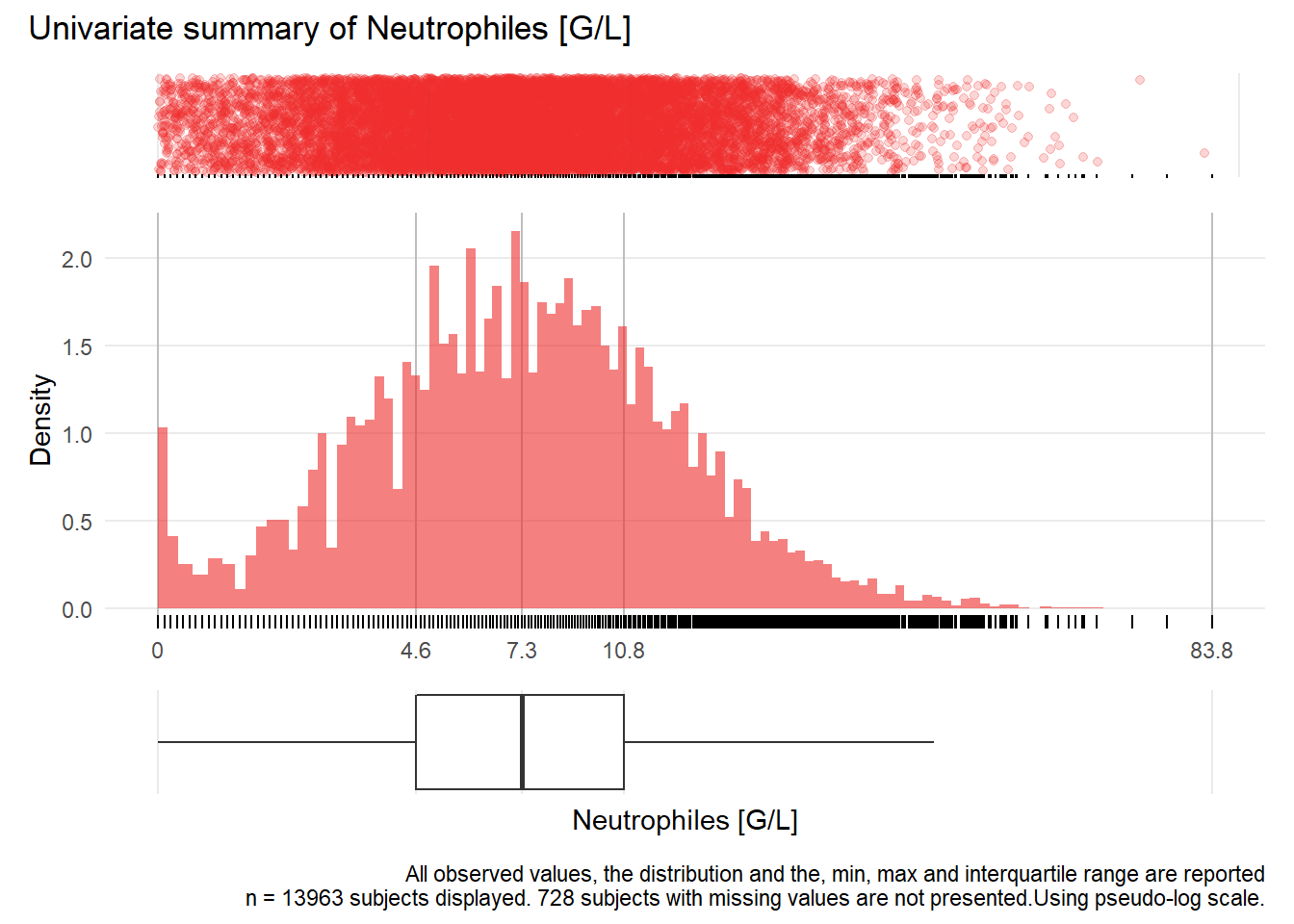
## Warning: Removed 1 rows containing missing values (geom_point).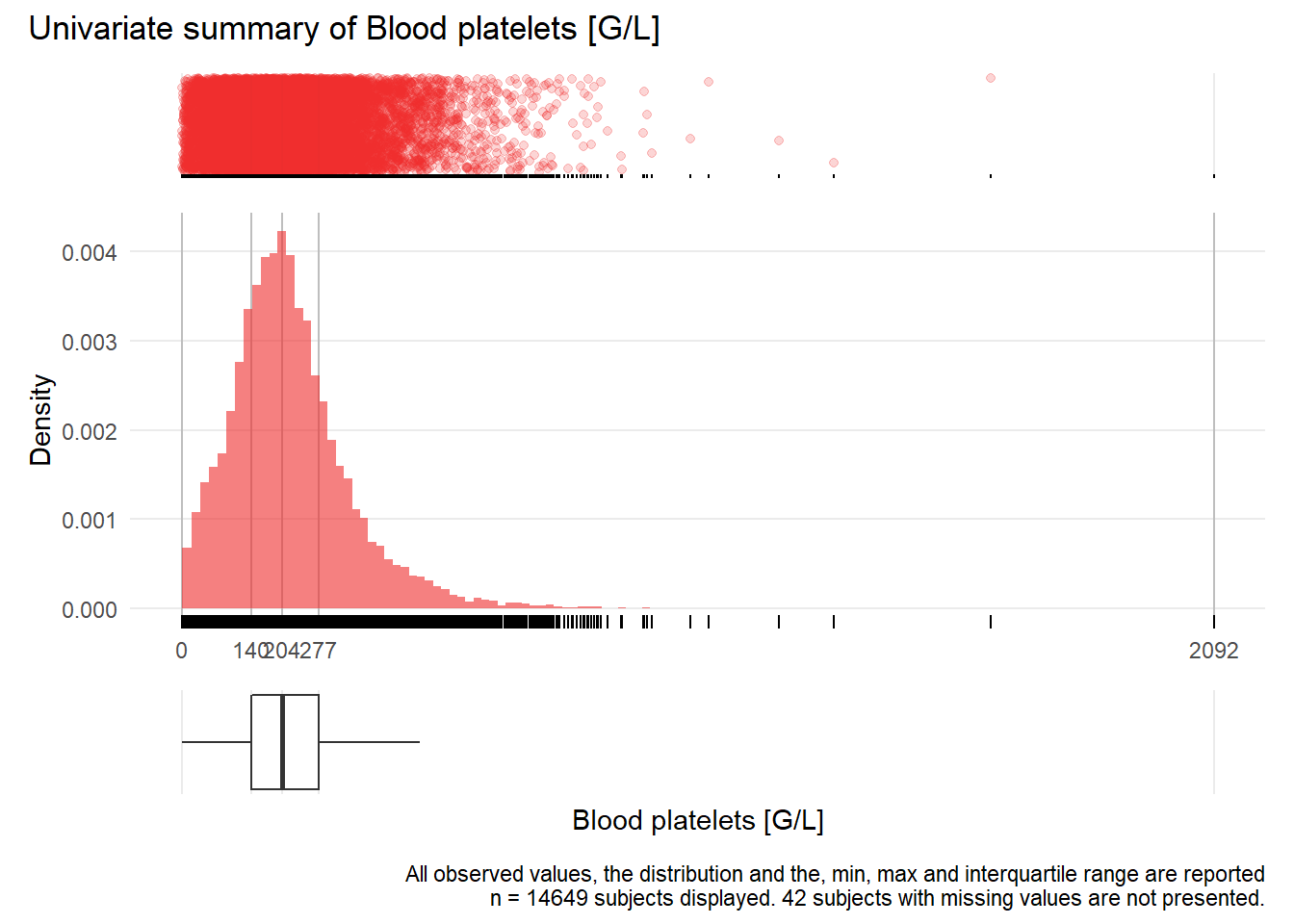
## Warning: Removed 3483 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).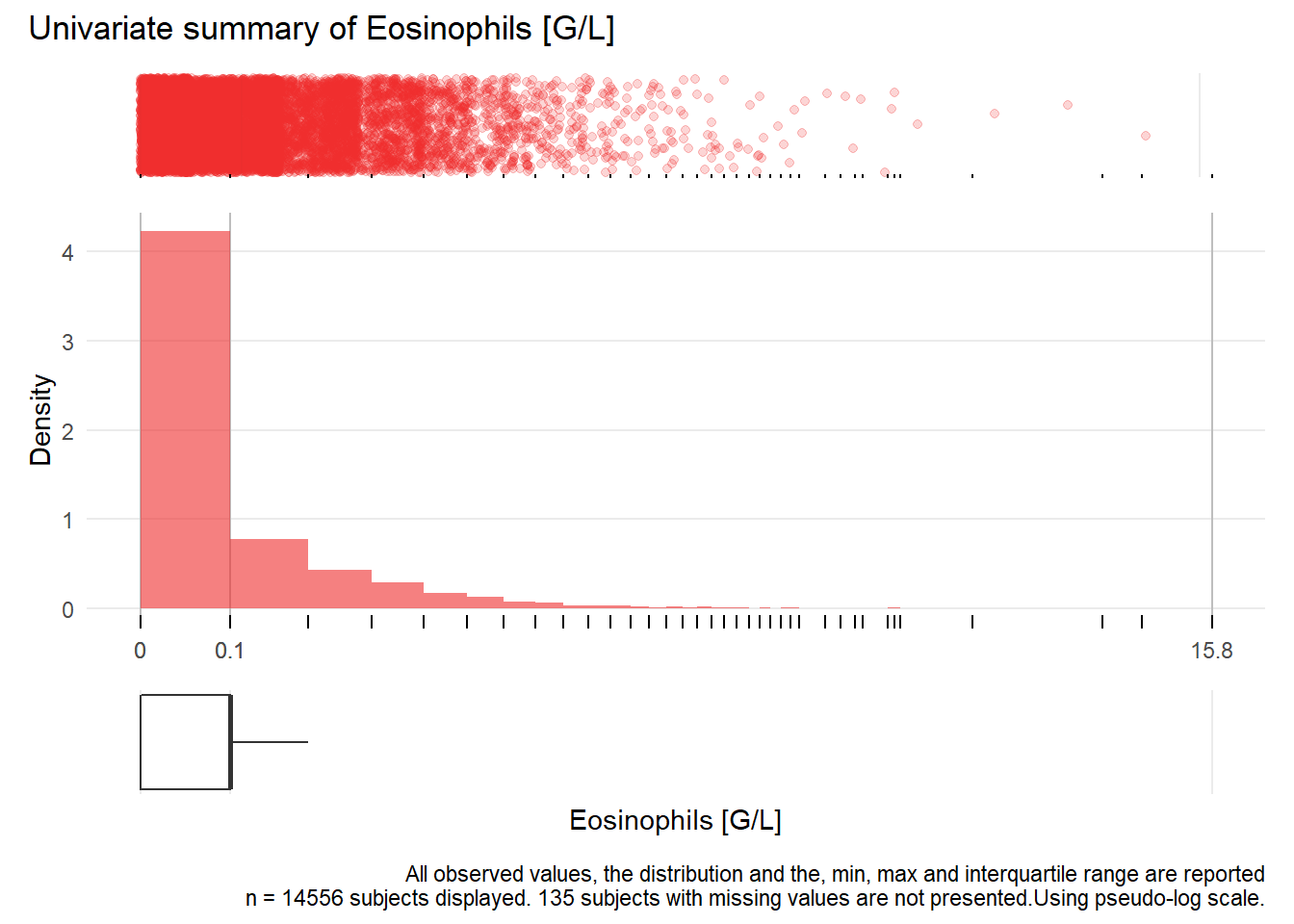
## Warning: Removed 6249 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).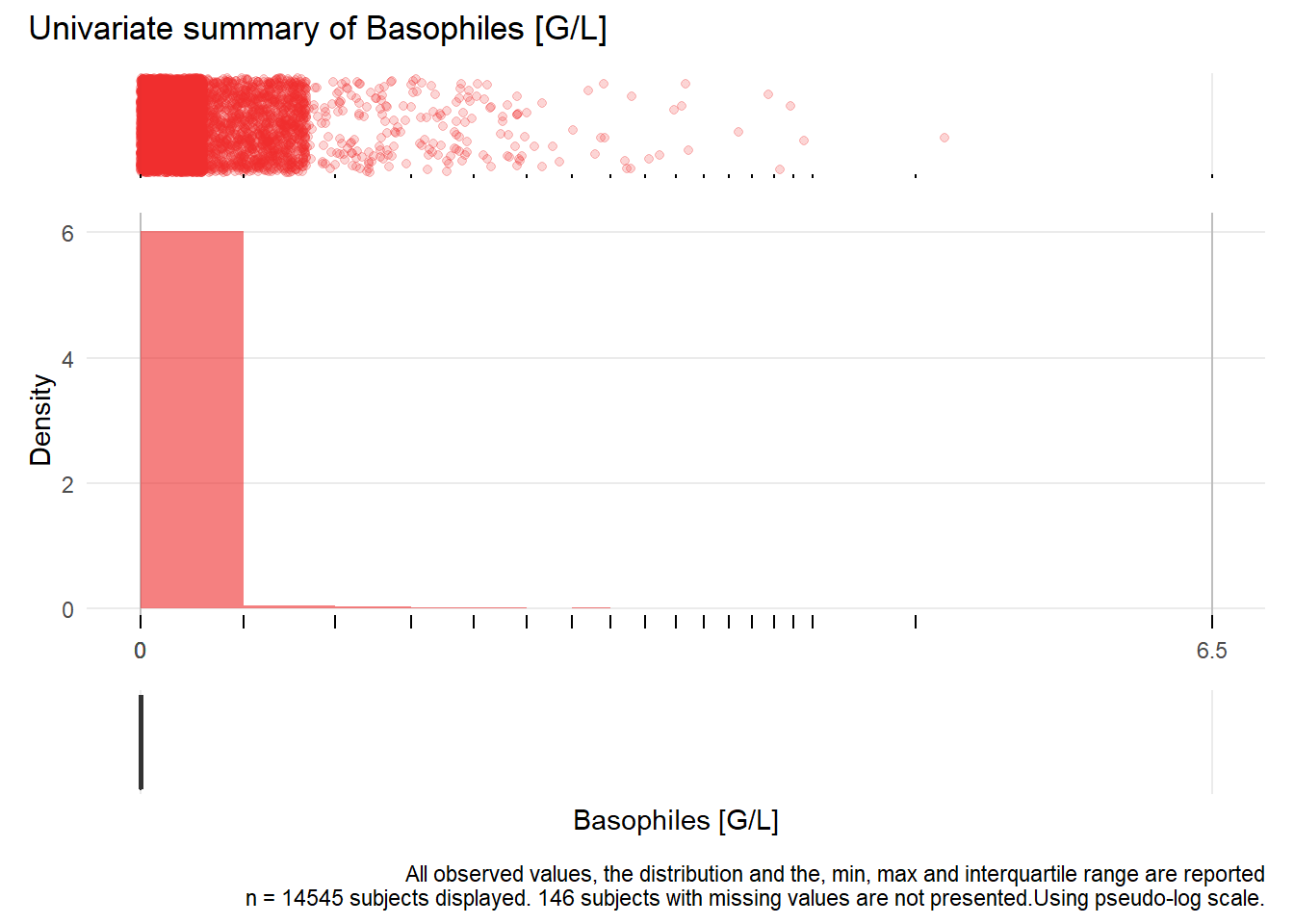
## Warning: Removed 56 rows containing missing values (geom_point).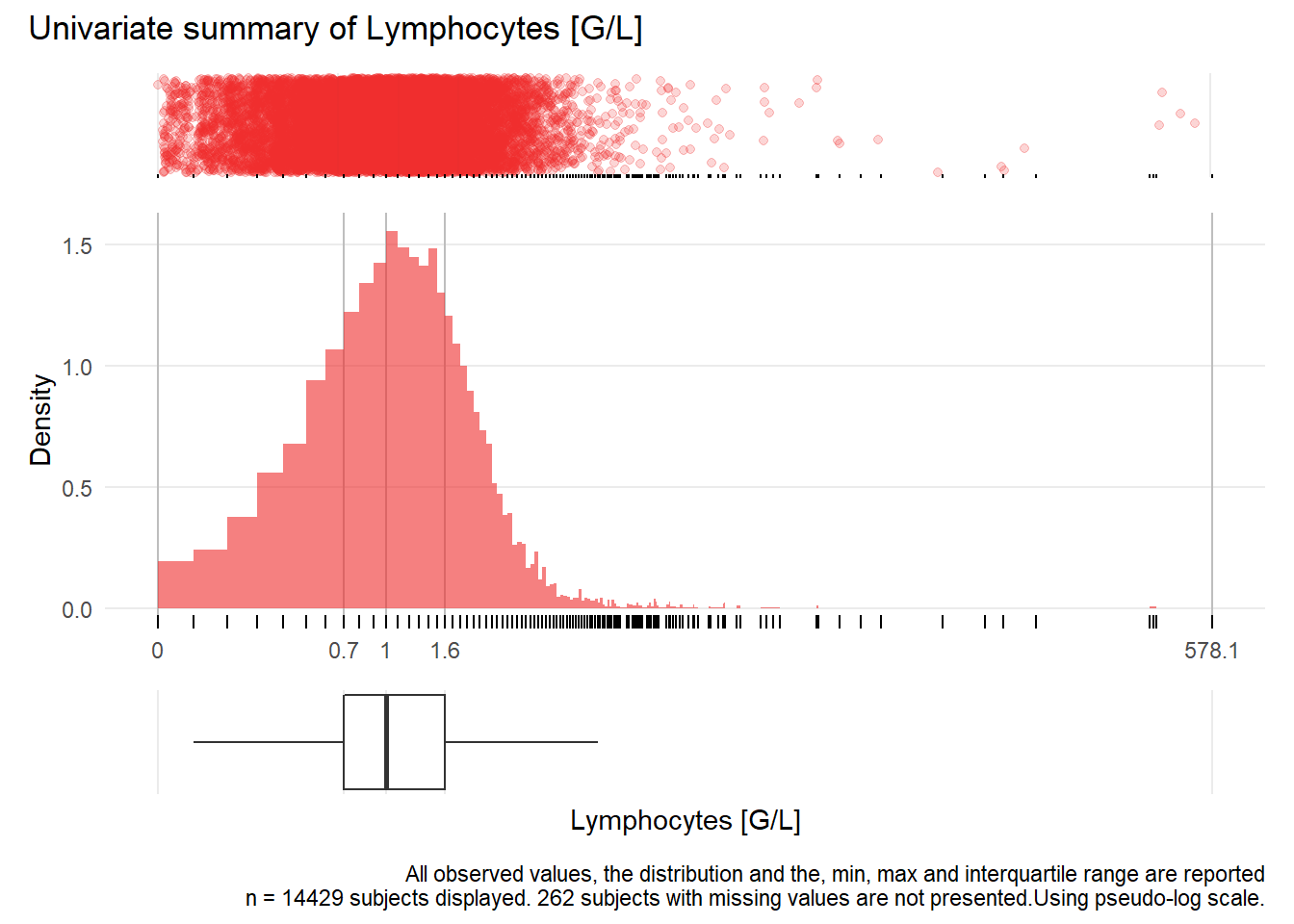
## Warning: Removed 233 rows containing missing values (geom_point).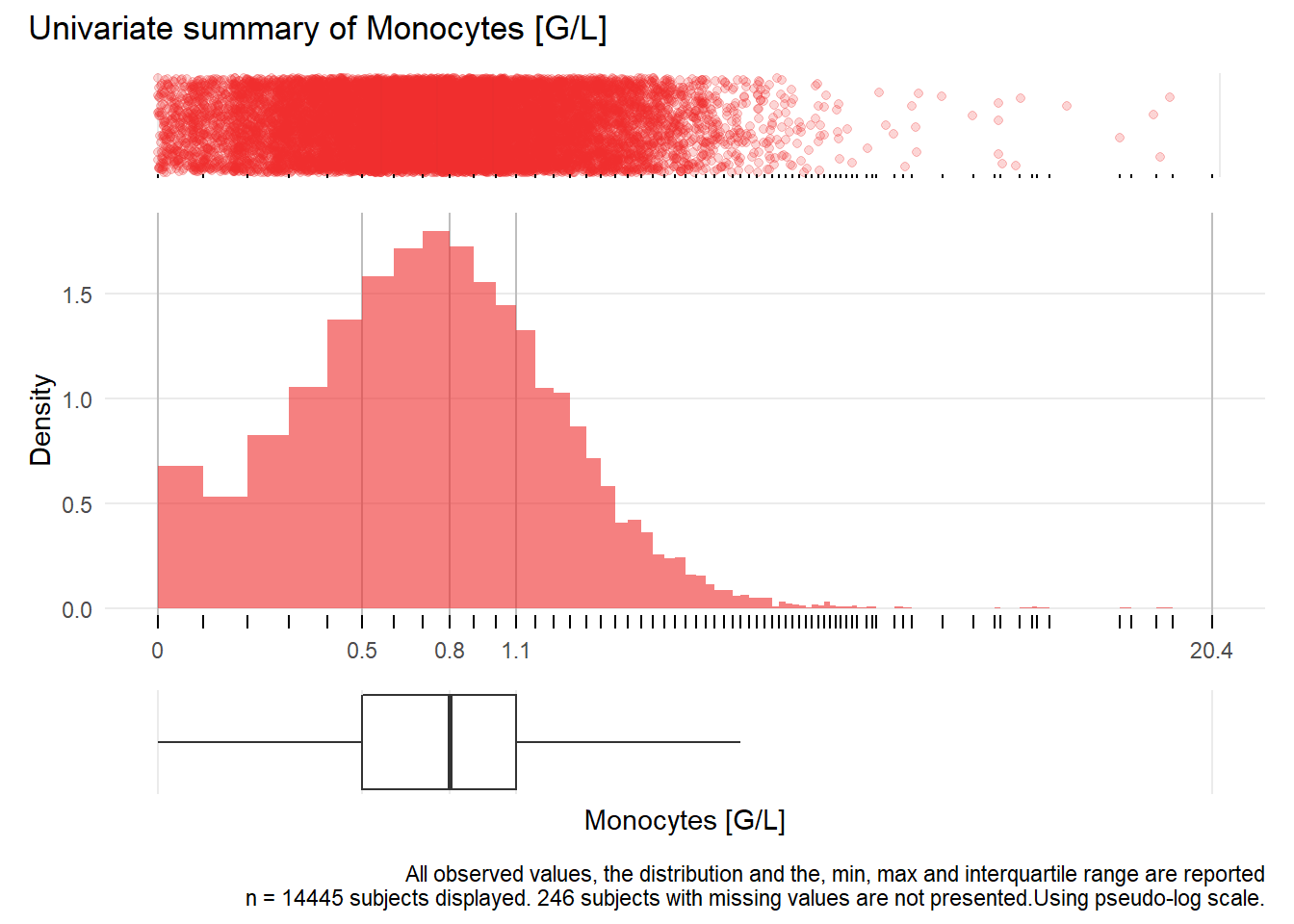
## Warning: Removed 76 rows containing missing values (geom_point).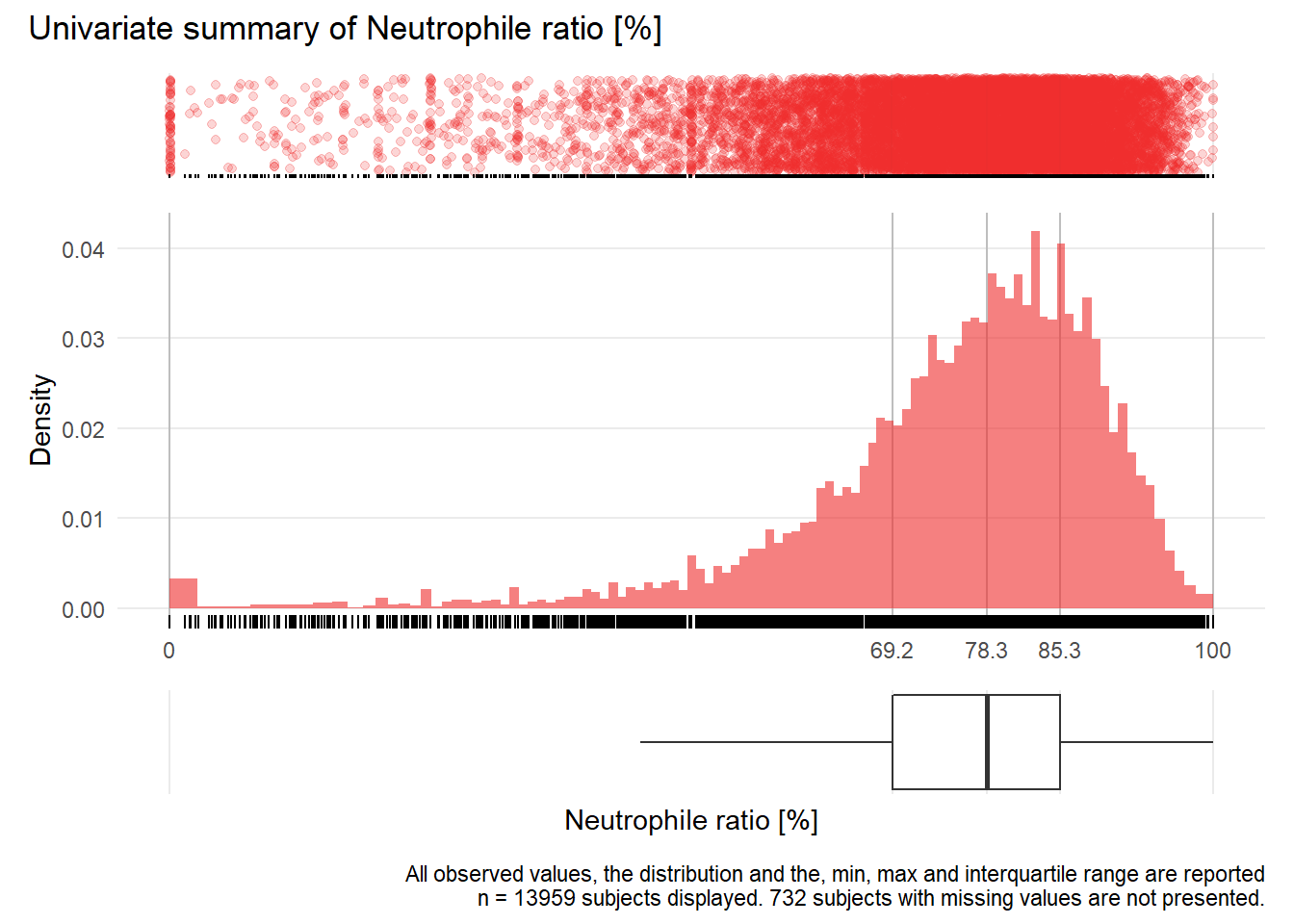
## Warning: Removed 3325 rows containing missing values (geom_point).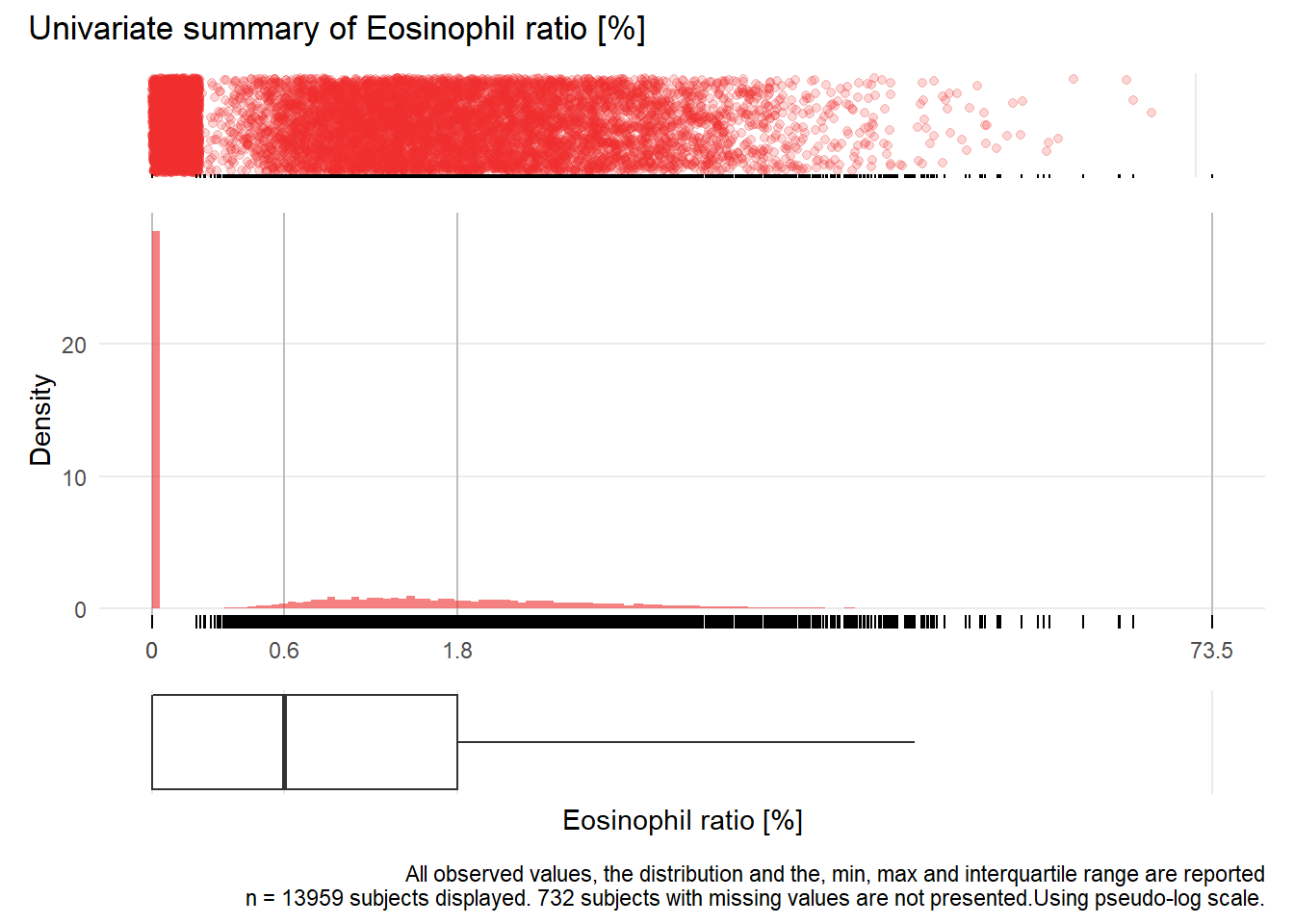
## Warning: Removed 6233 rows containing missing values (geom_point).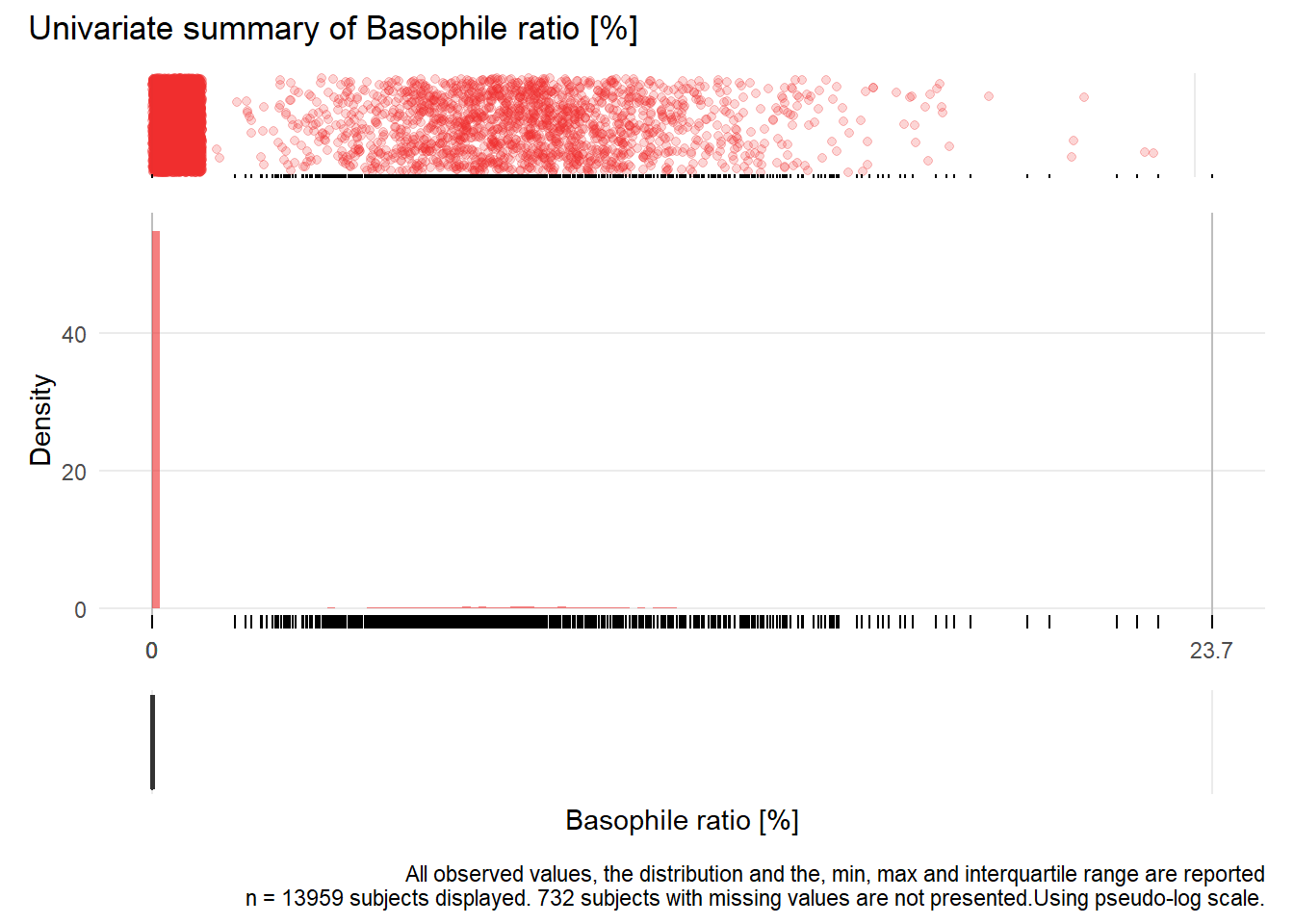
## Warning: Removed 92 rows containing missing values (geom_point).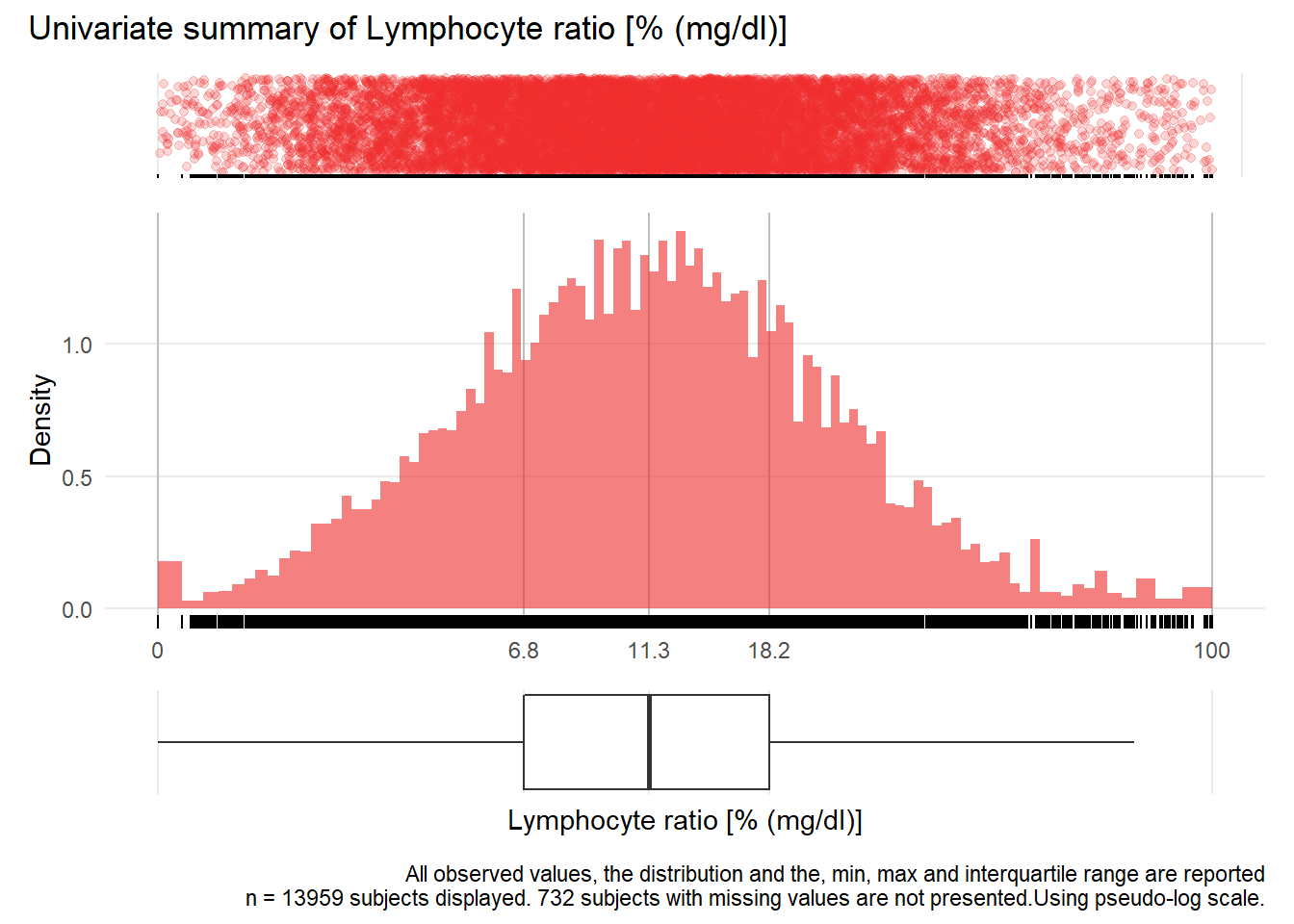
## Warning: Removed 204 rows containing missing values (geom_point).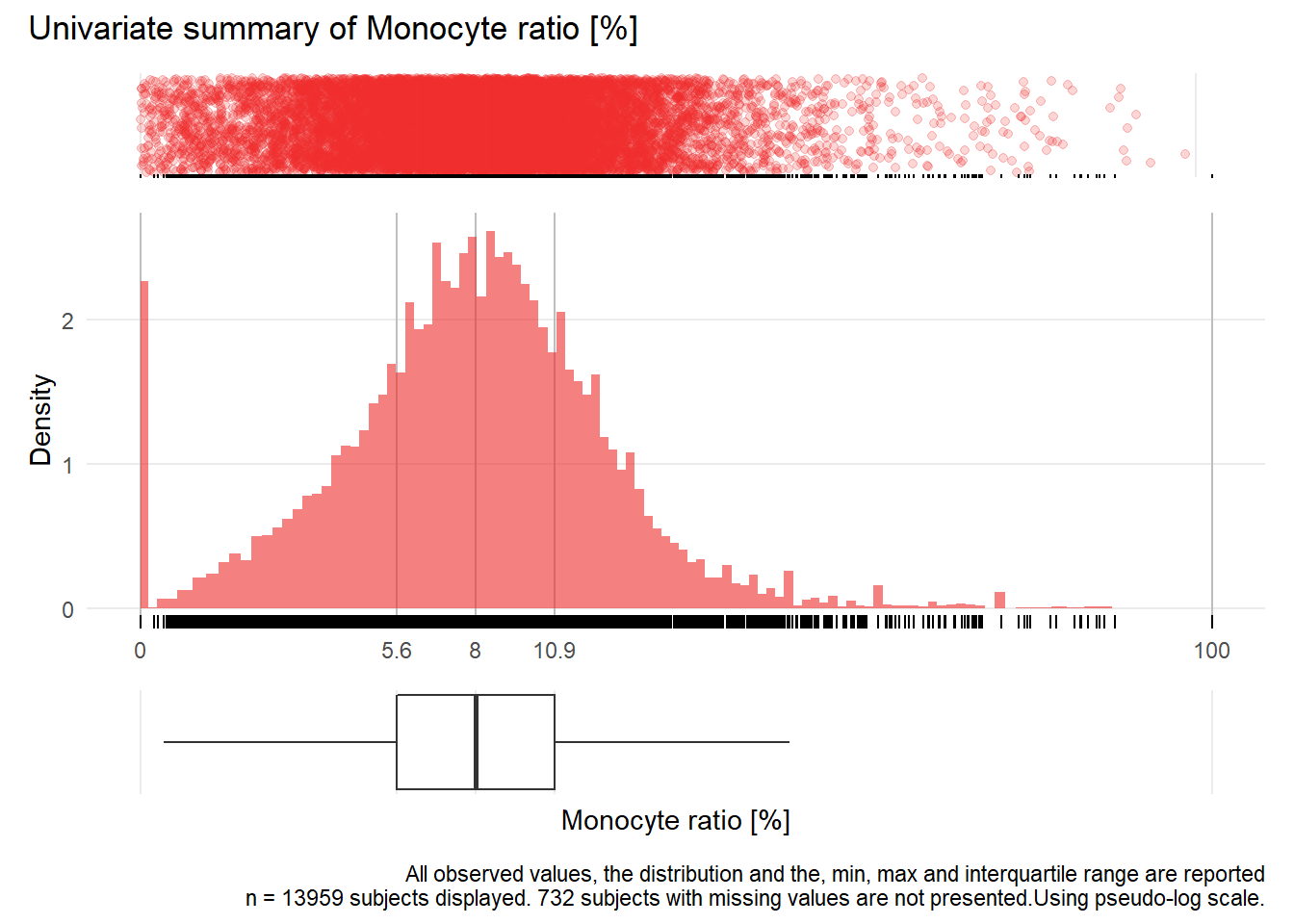
## Warning: Removed 6 rows containing missing values (geom_point).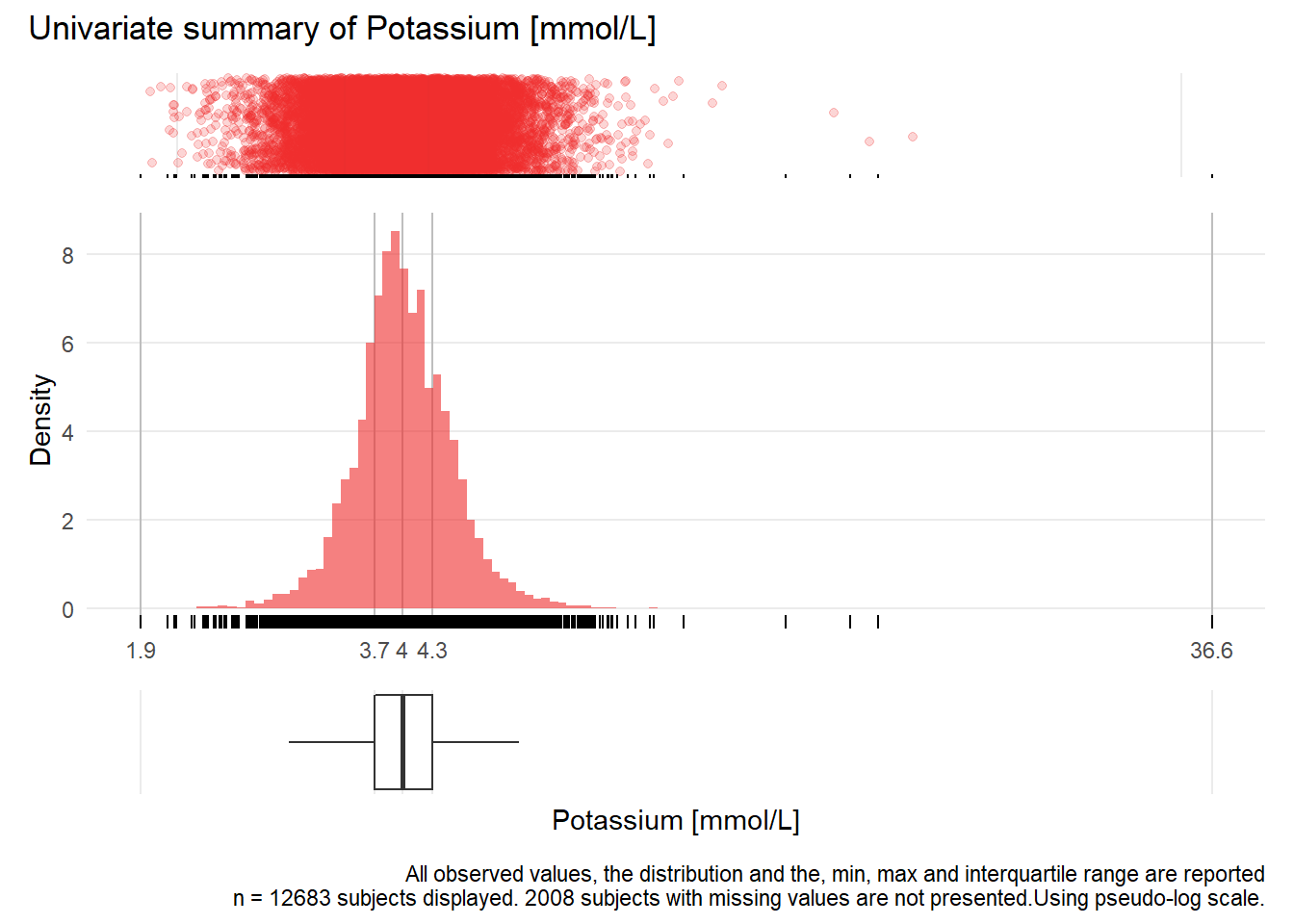
## Warning: Removed 2 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).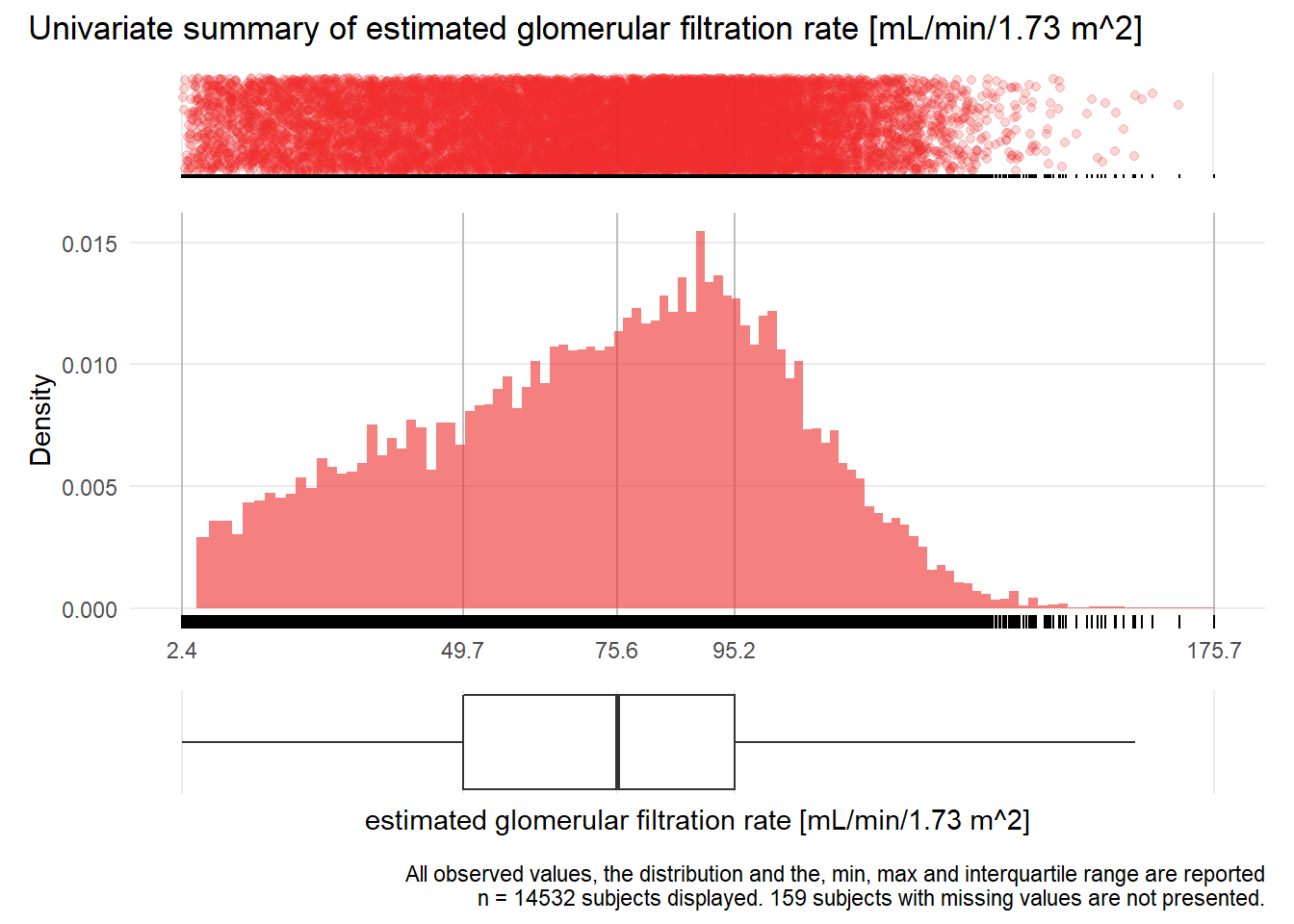
## Warning: Removed 1 rows containing missing values (geom_point).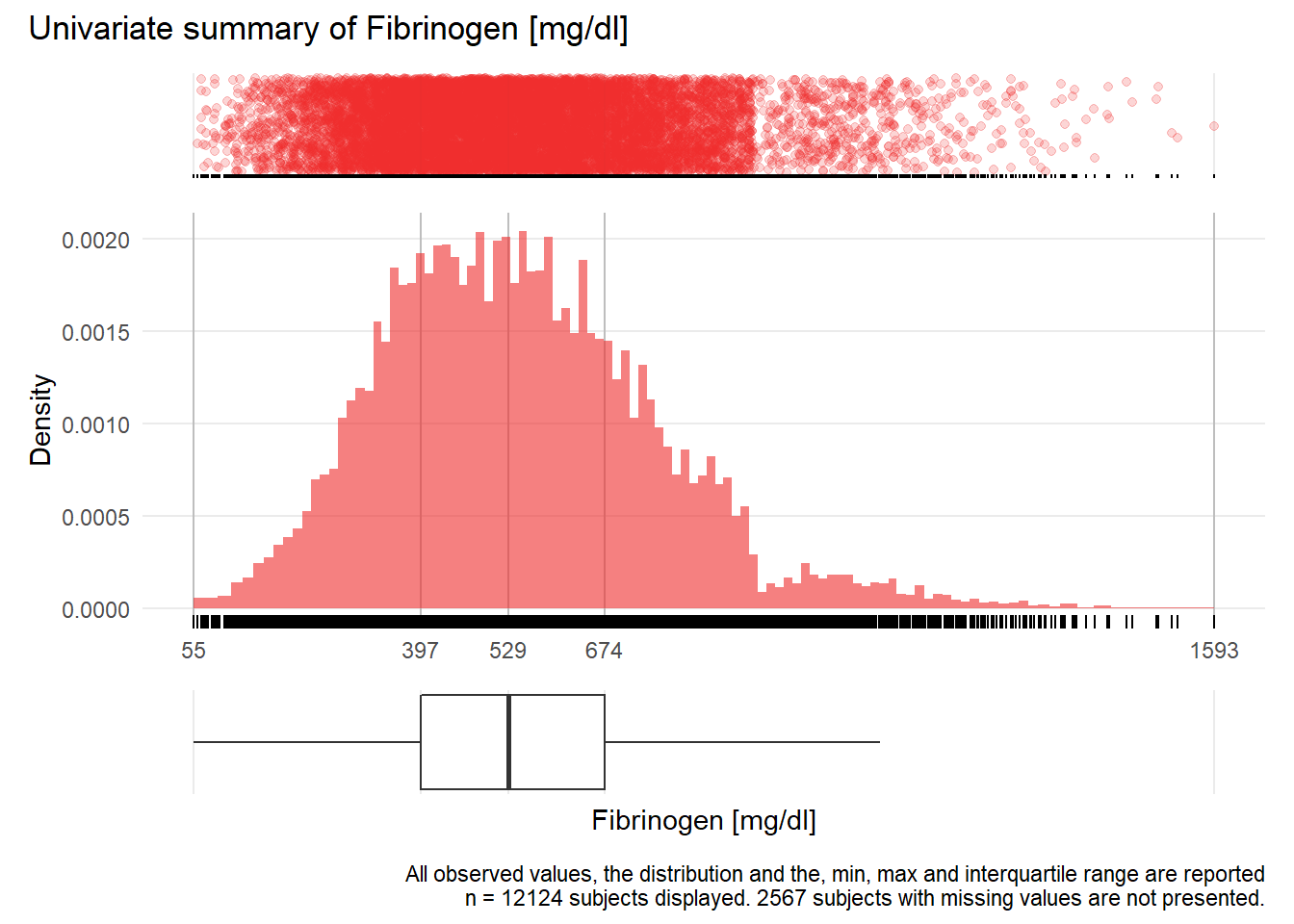
## Warning: Removed 57 rows containing missing values (geom_point).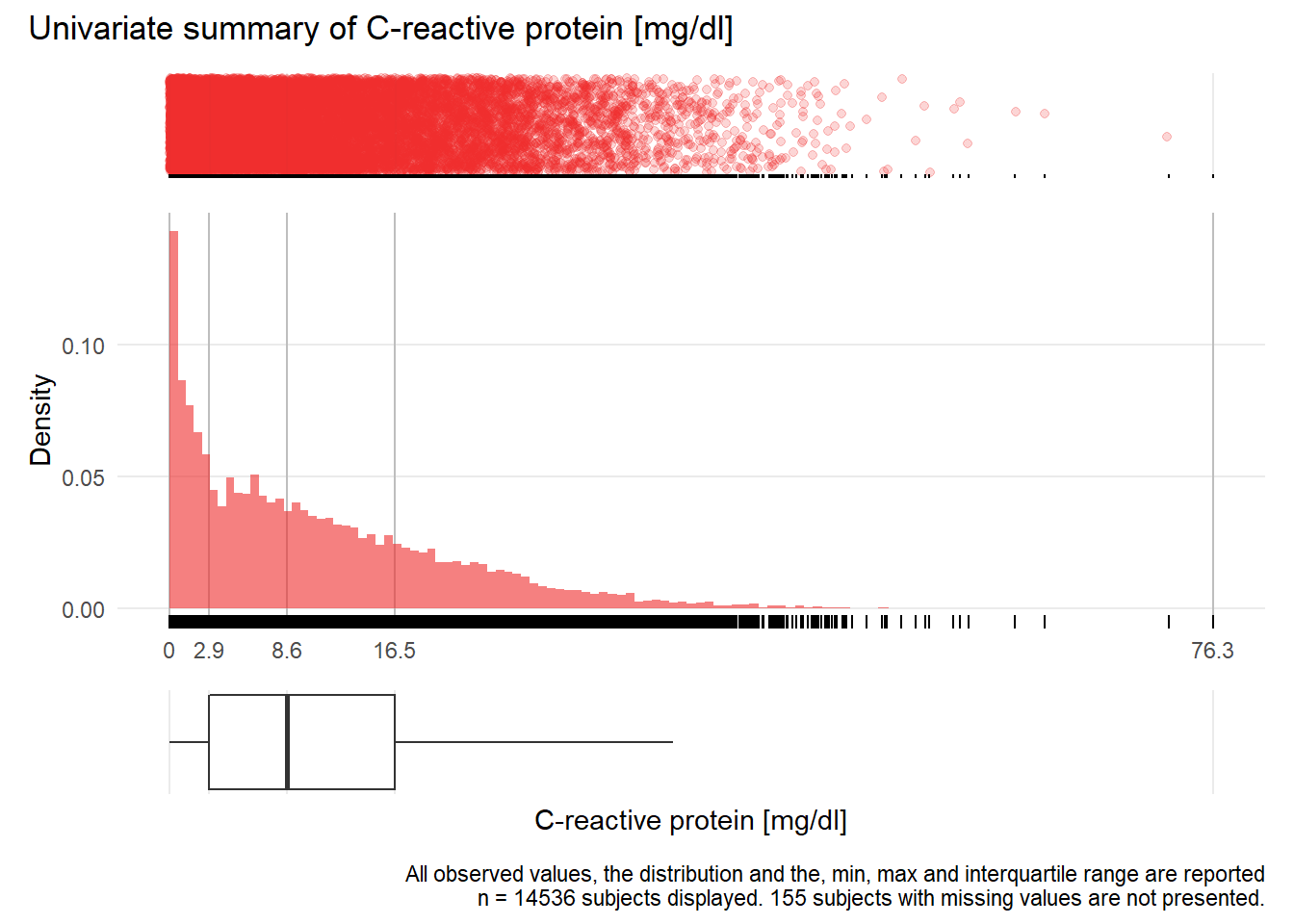
## Warning: Removed 1 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).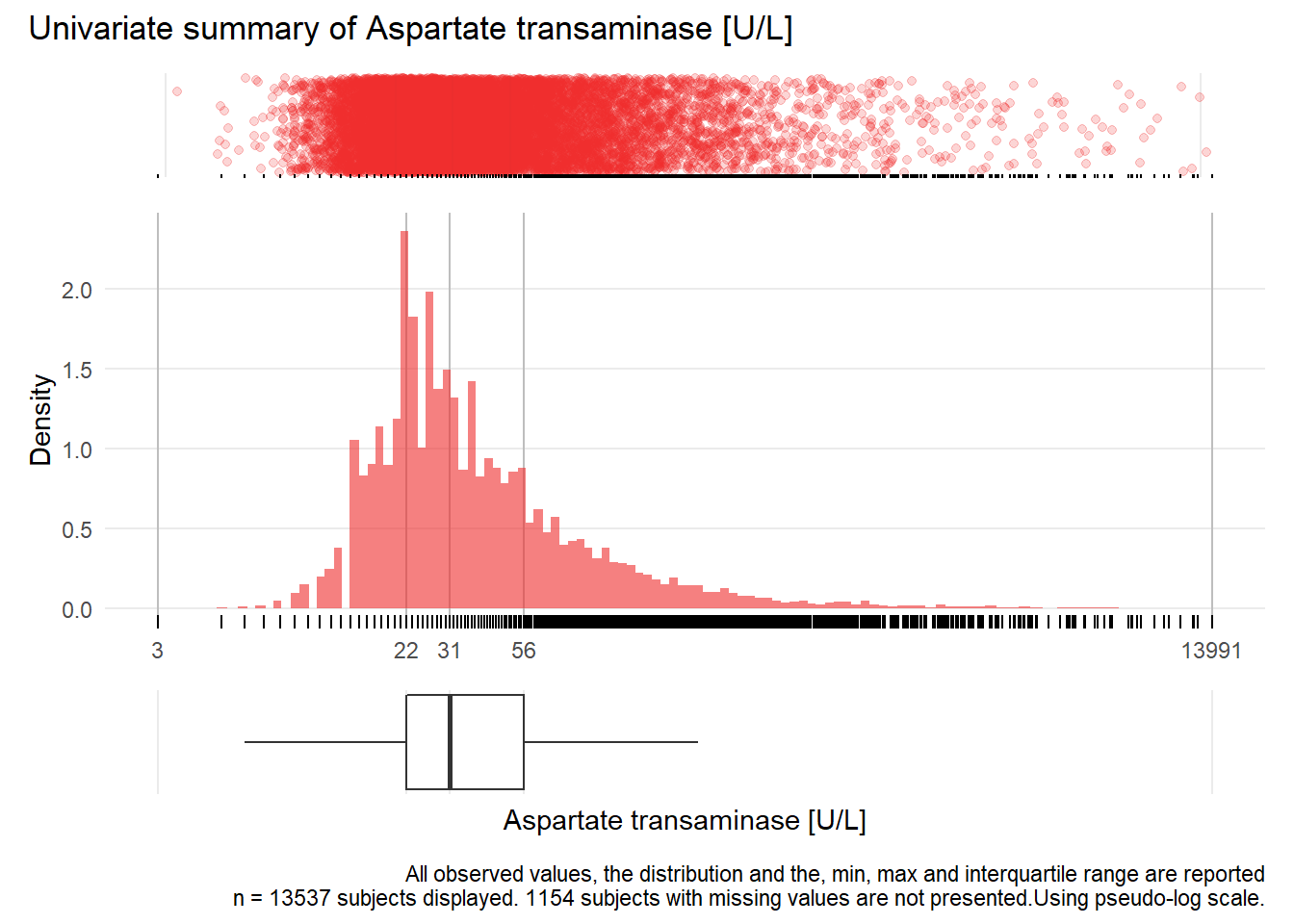
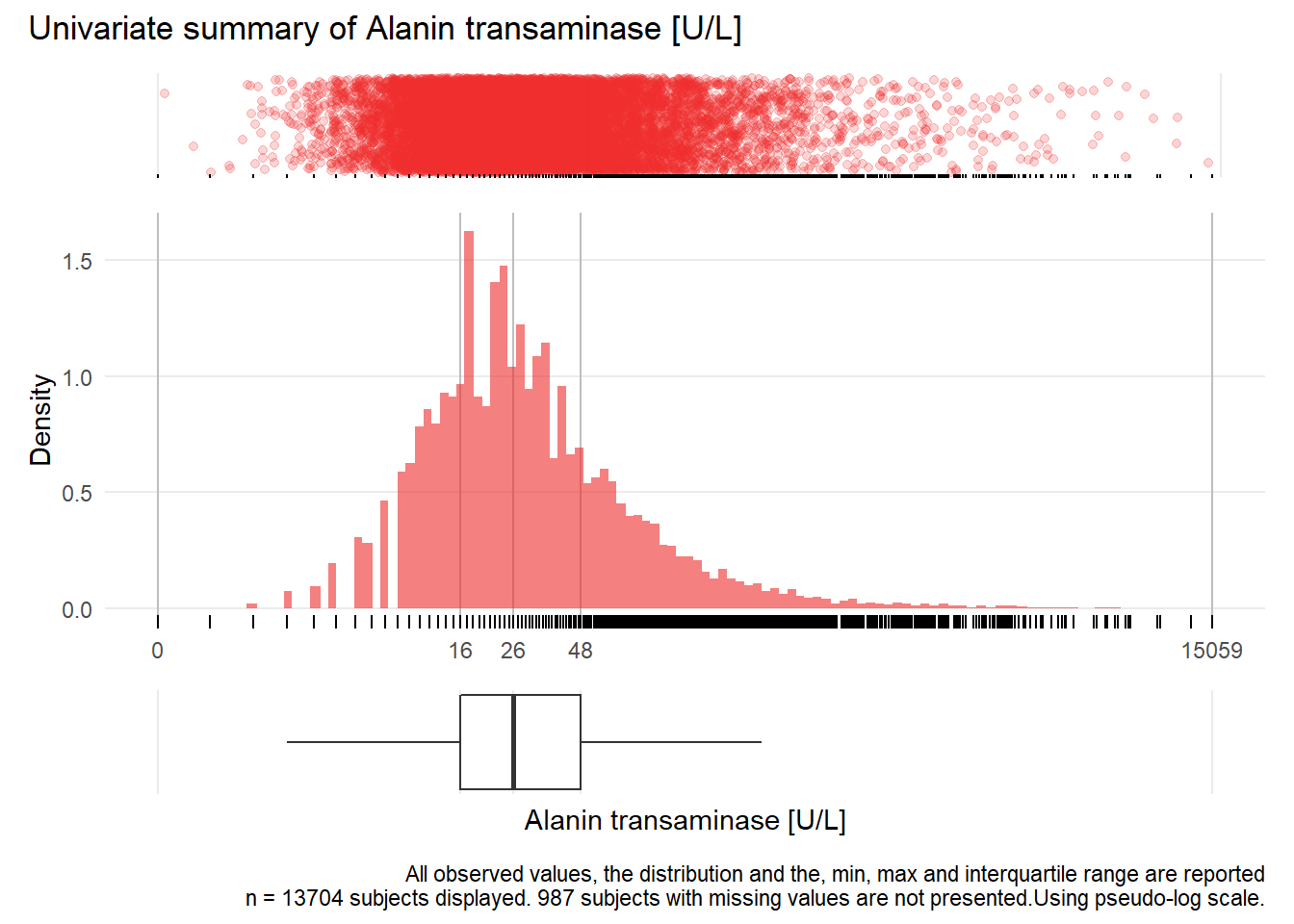
## Warning: Removed 2 rows containing missing values (geom_point).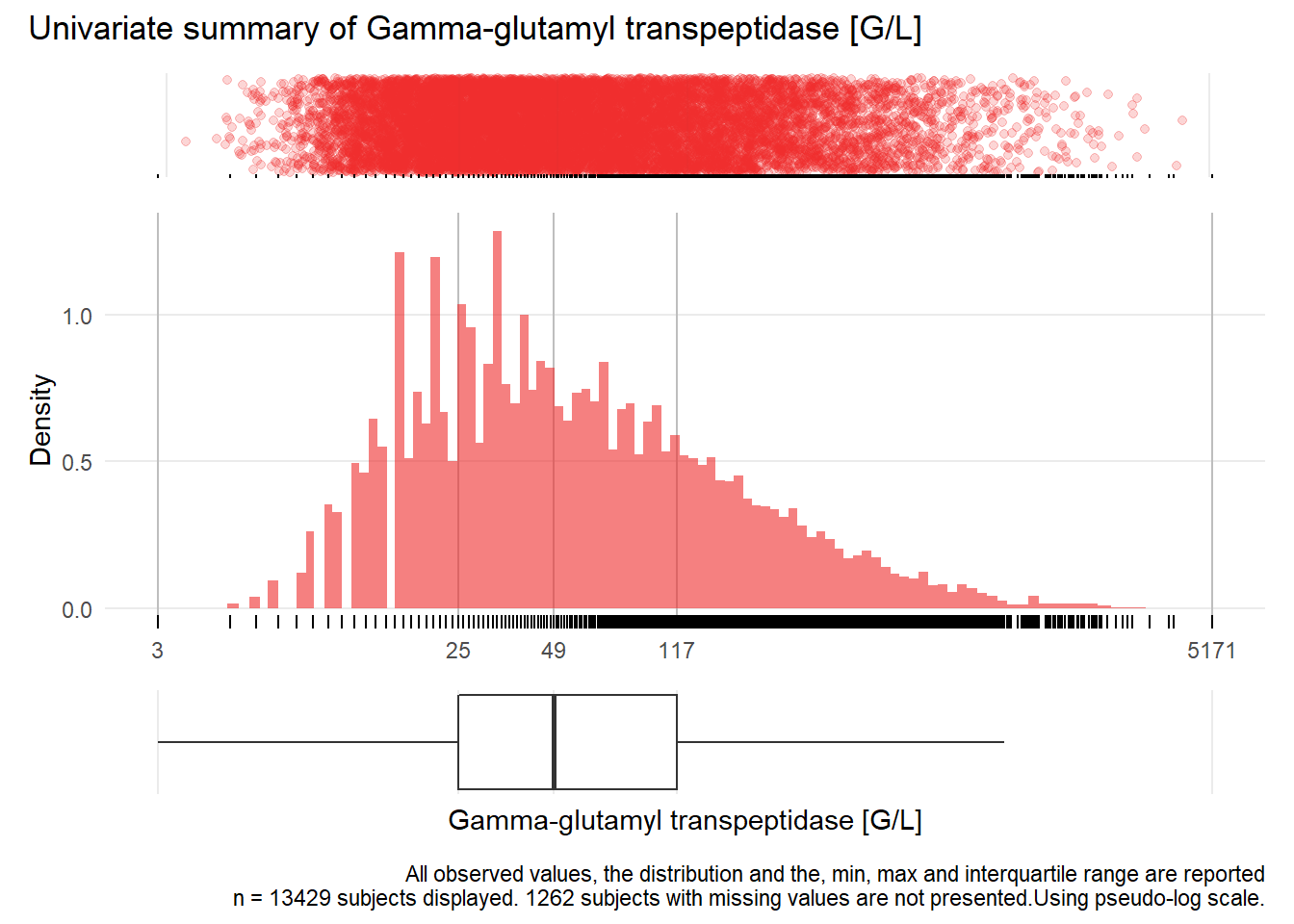
## Warning: Removed 1 rows containing missing values (geom_point).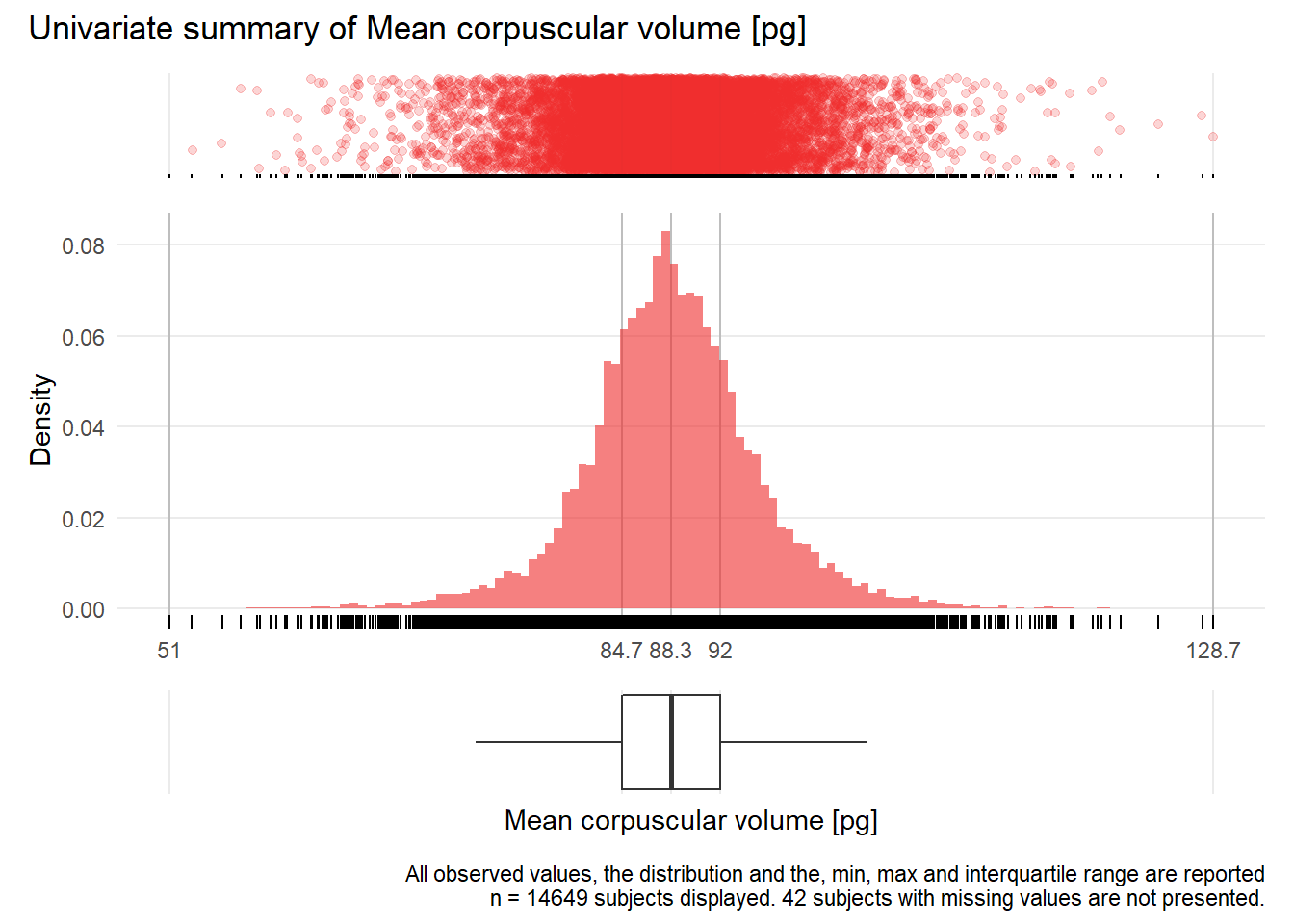
## Warning: Removed 7 rows containing missing values (geom_point).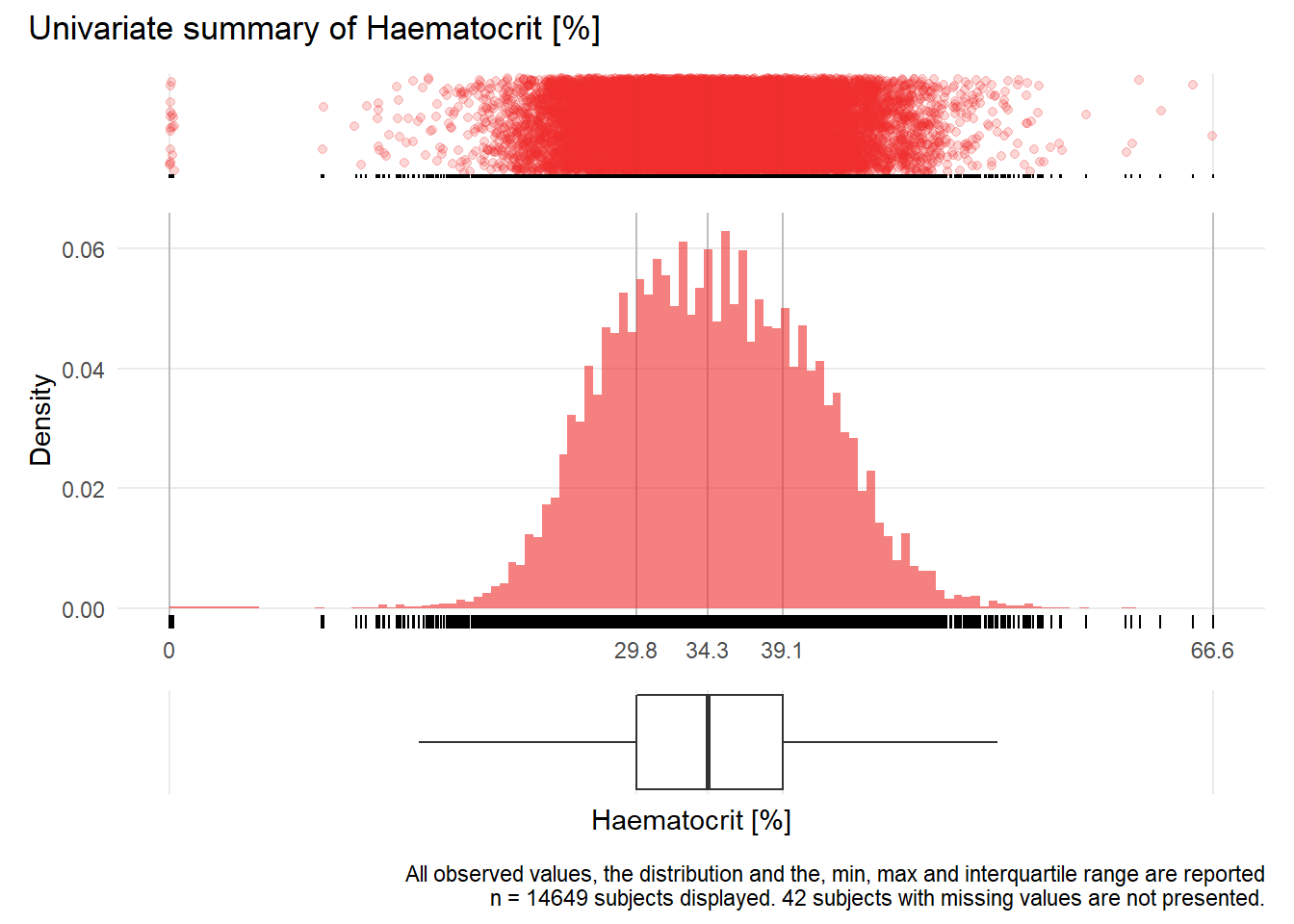
## Warning: Removed 2 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).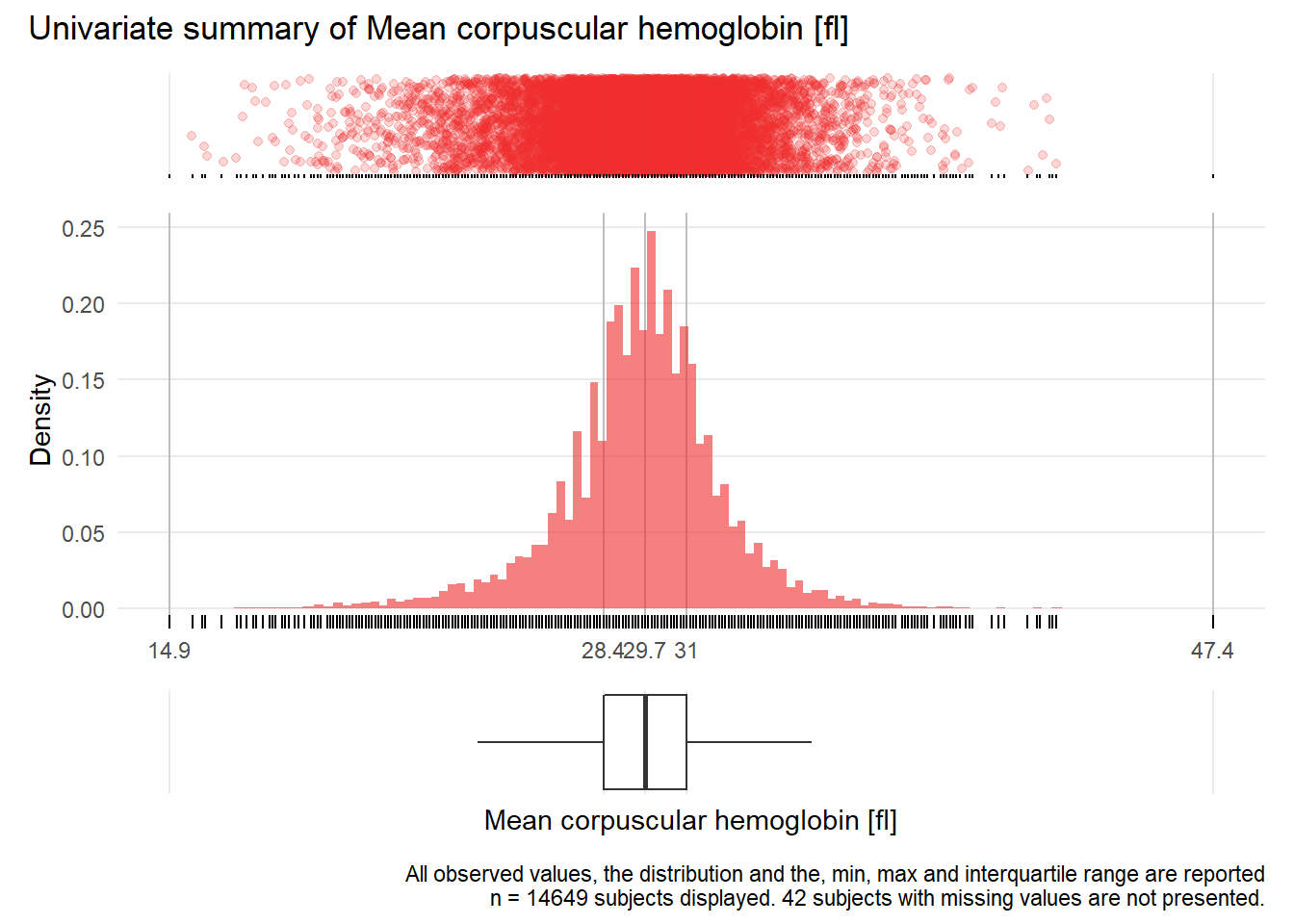
## Warning: Removed 1 rows containing missing values (geom_bar).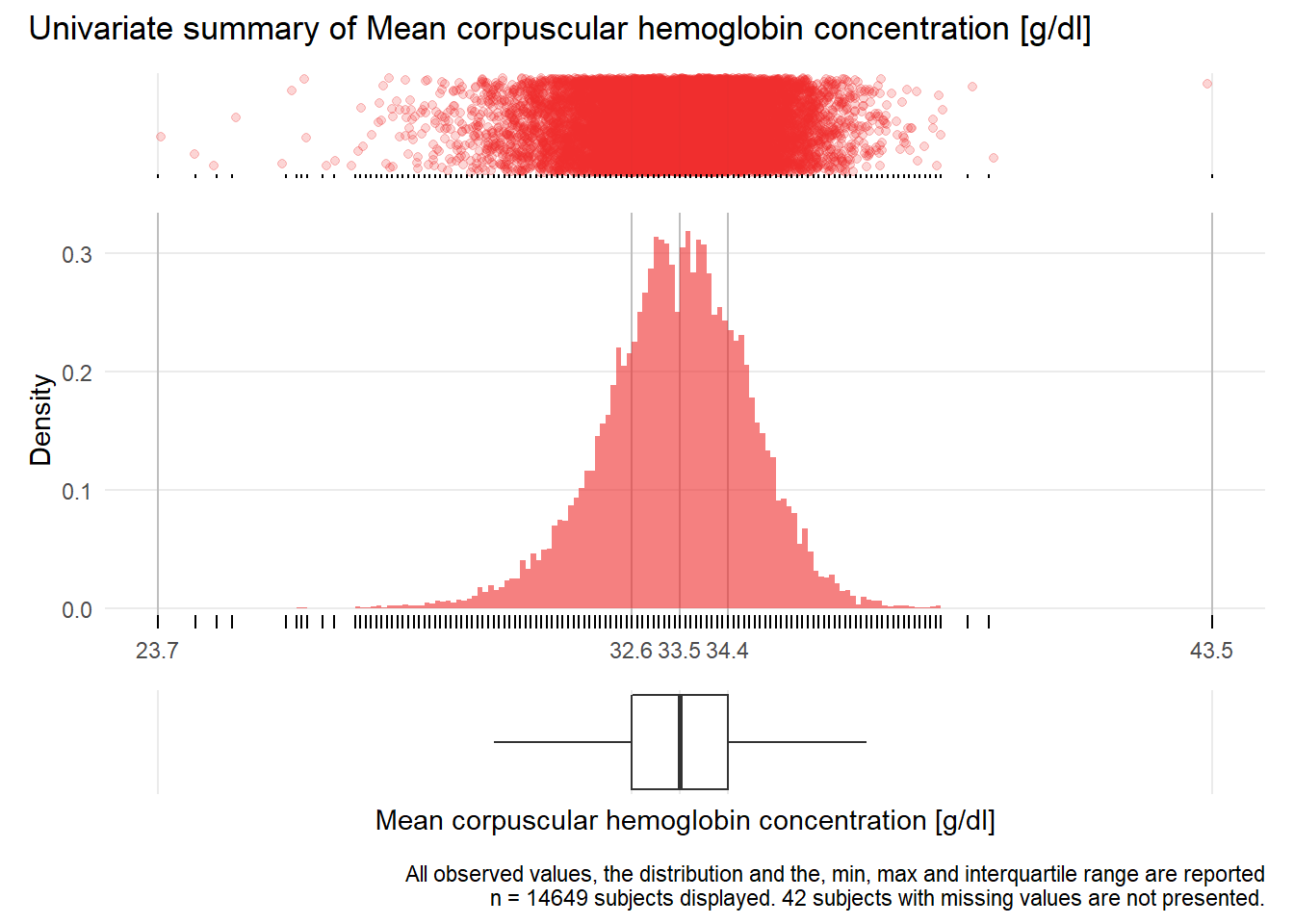
## Warning: Removed 1 rows containing missing values (geom_point).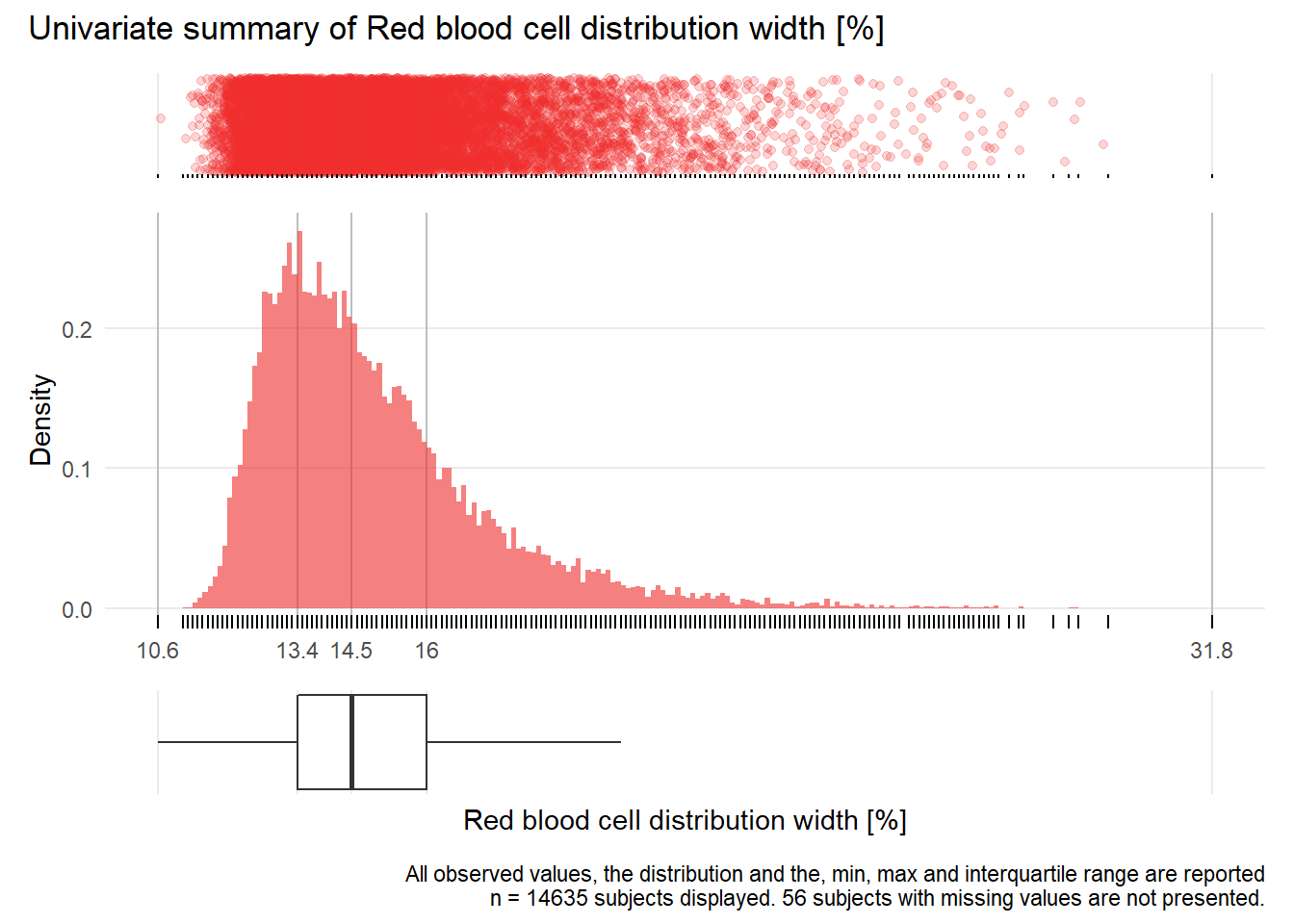
## Warning: Removed 1 rows containing missing values (geom_point).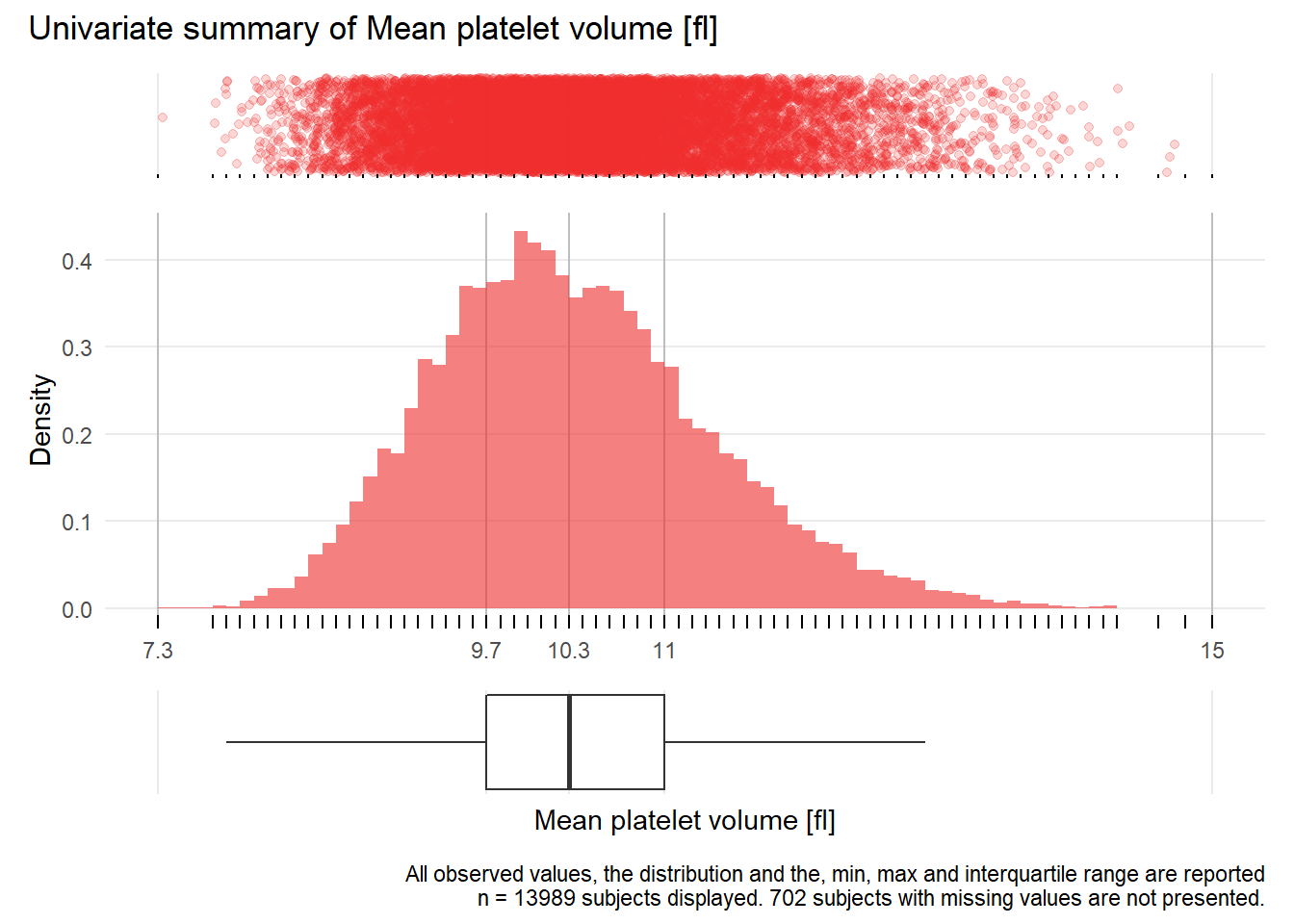
## Warning: Removed 2 rows containing missing values (geom_point).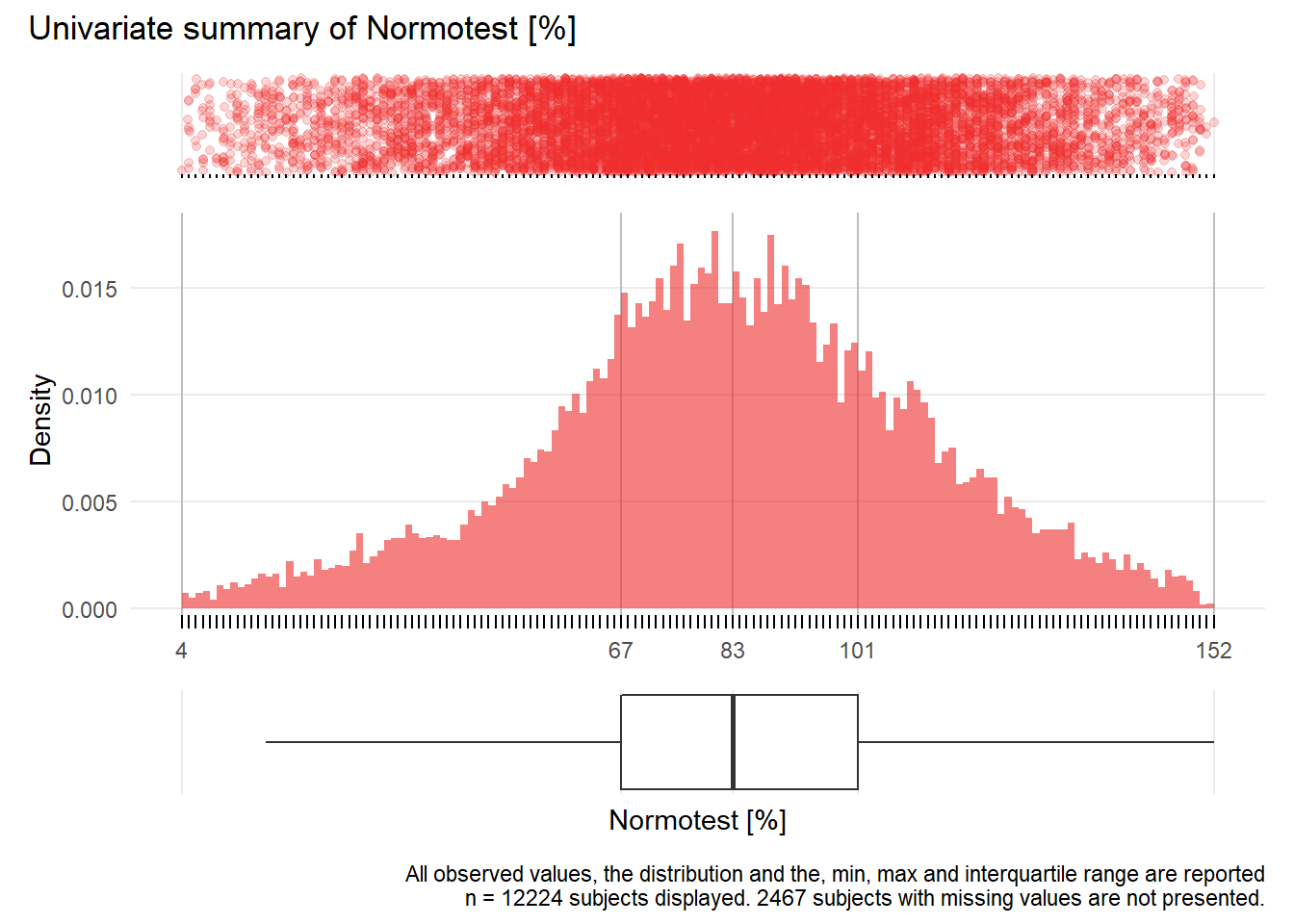
## Warning: Removed 12 rows containing missing values (geom_point).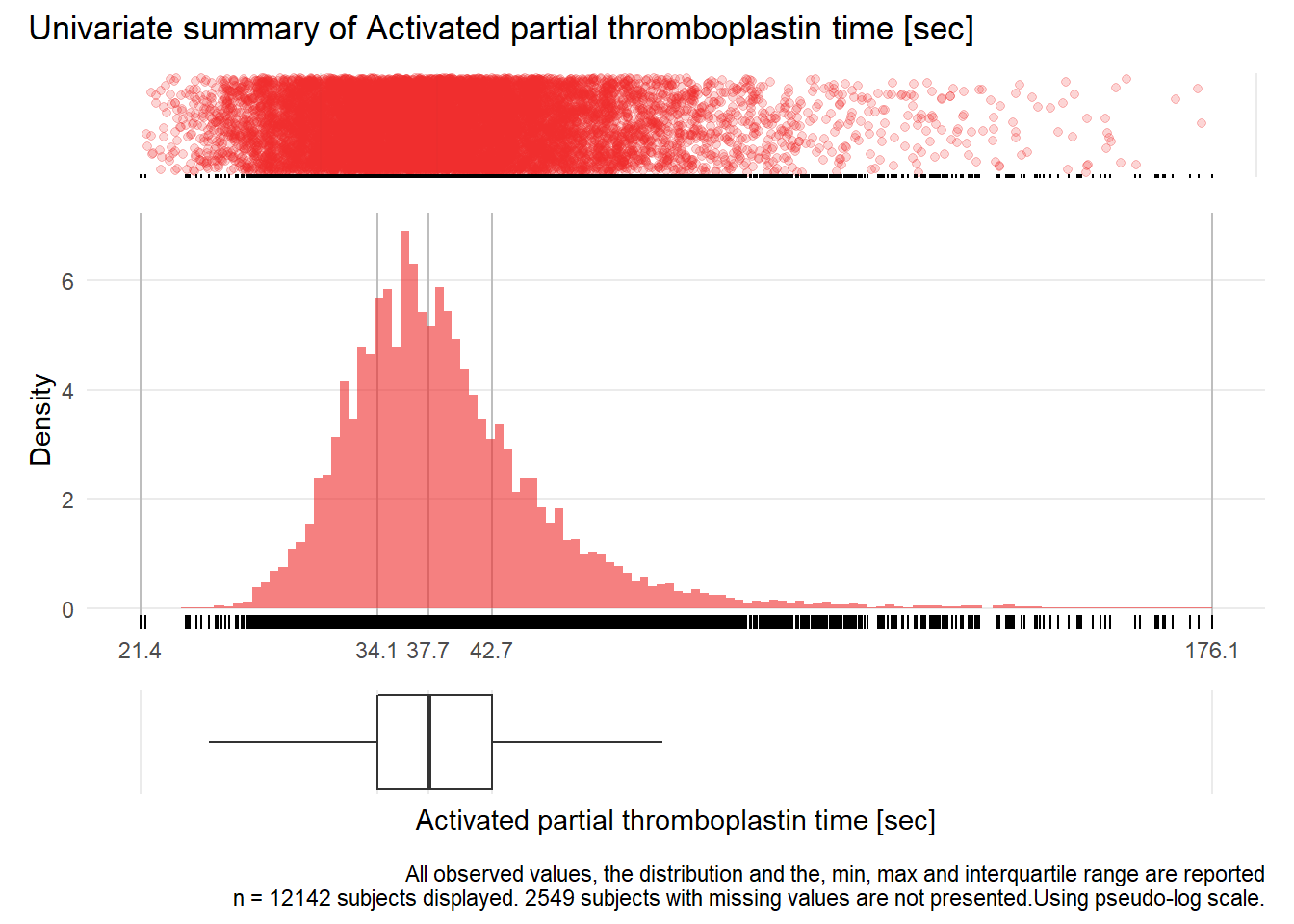
## Warning: Removed 1 rows containing missing values (geom_point).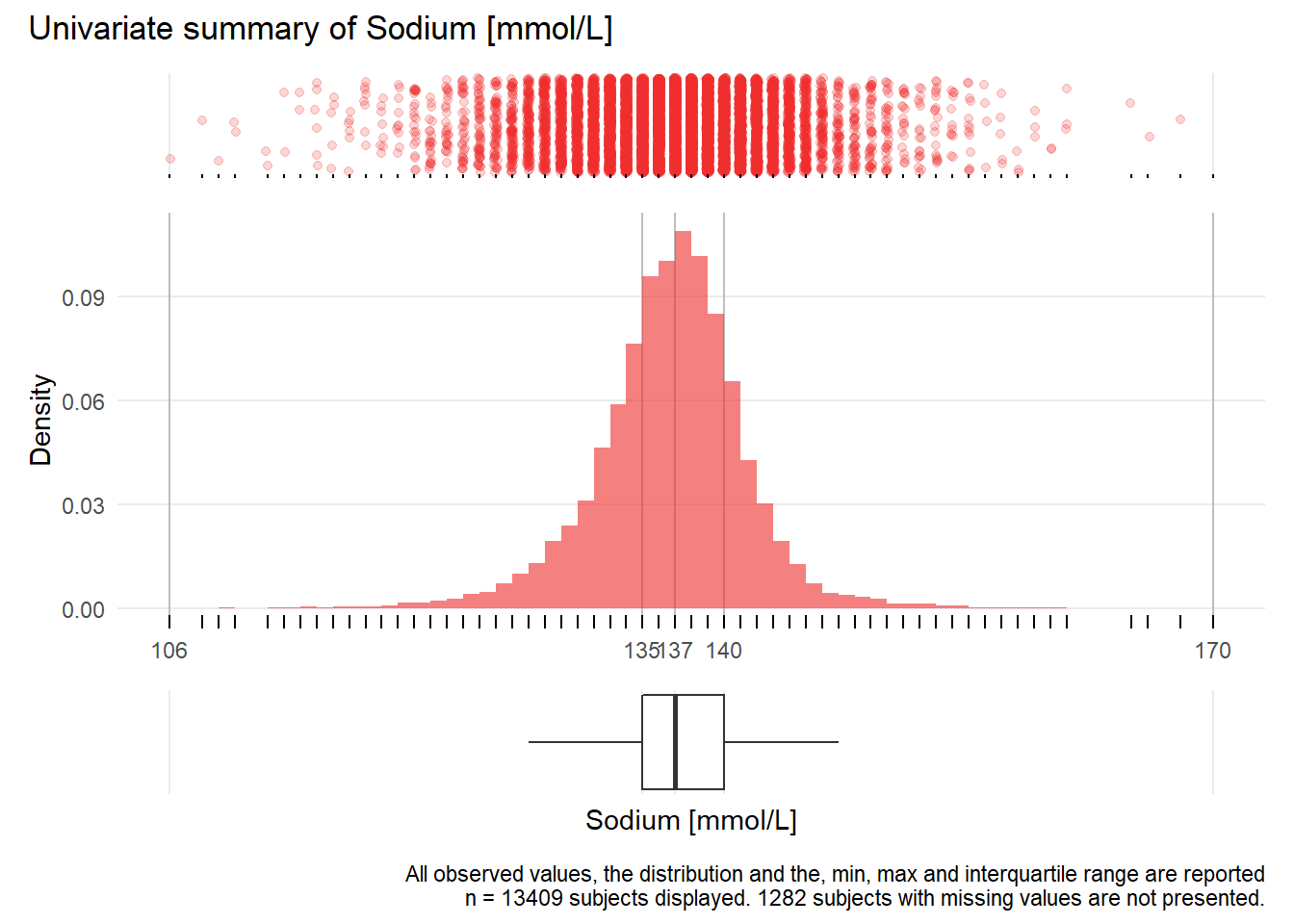
## Warning: Removed 2 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).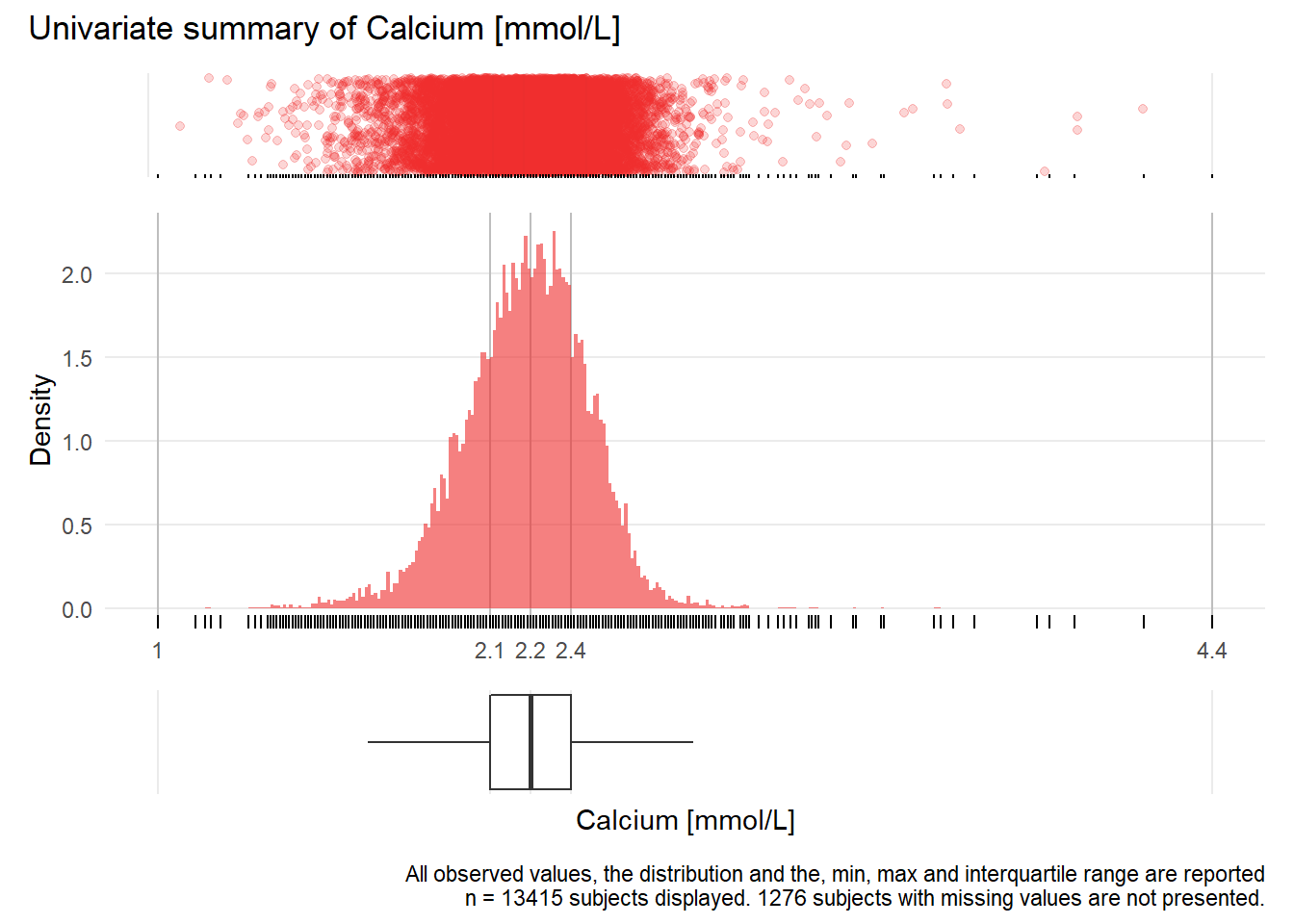
## Warning: Removed 27 rows containing missing values (geom_point).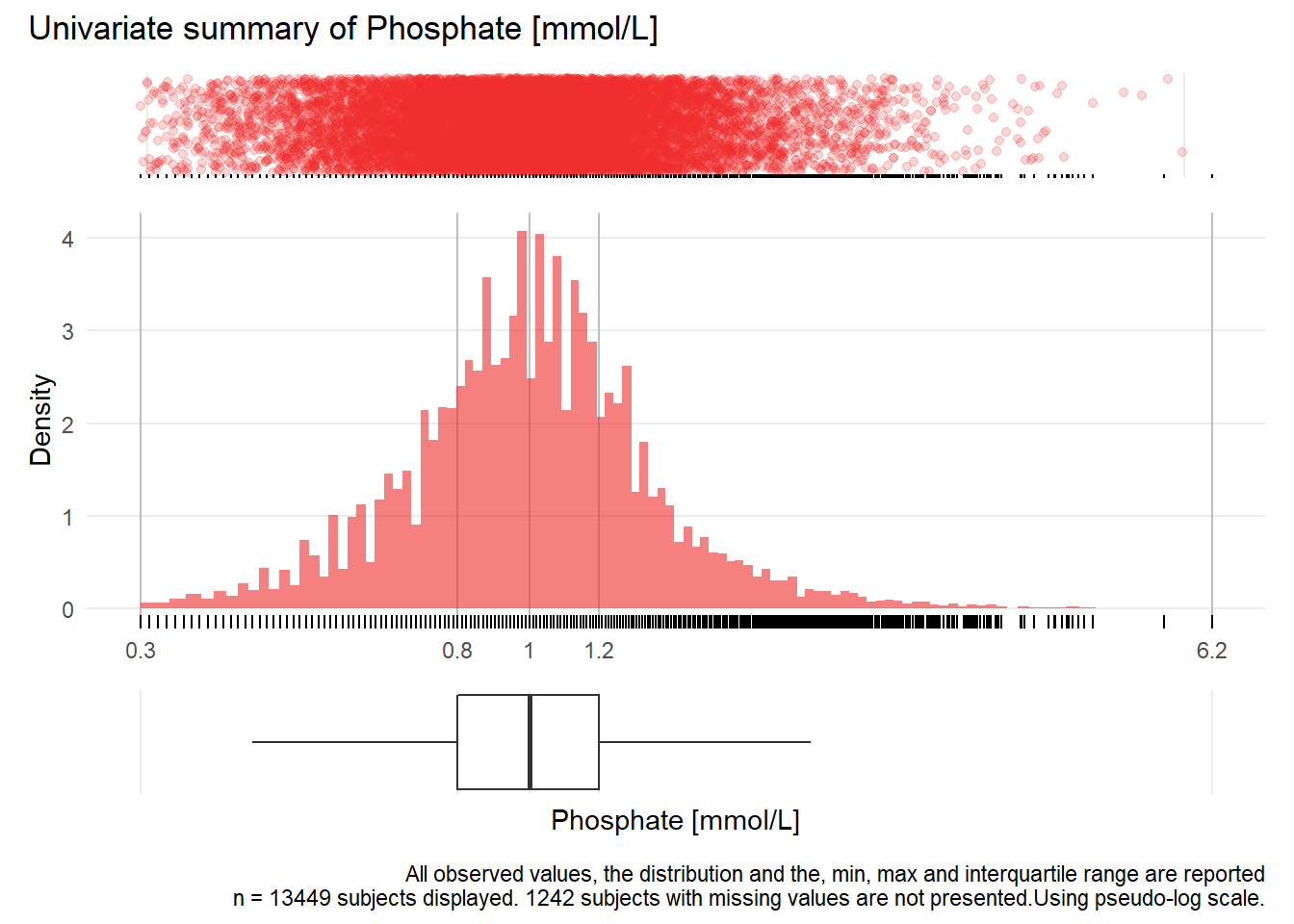
## Warning: Removed 5 rows containing missing values (geom_point).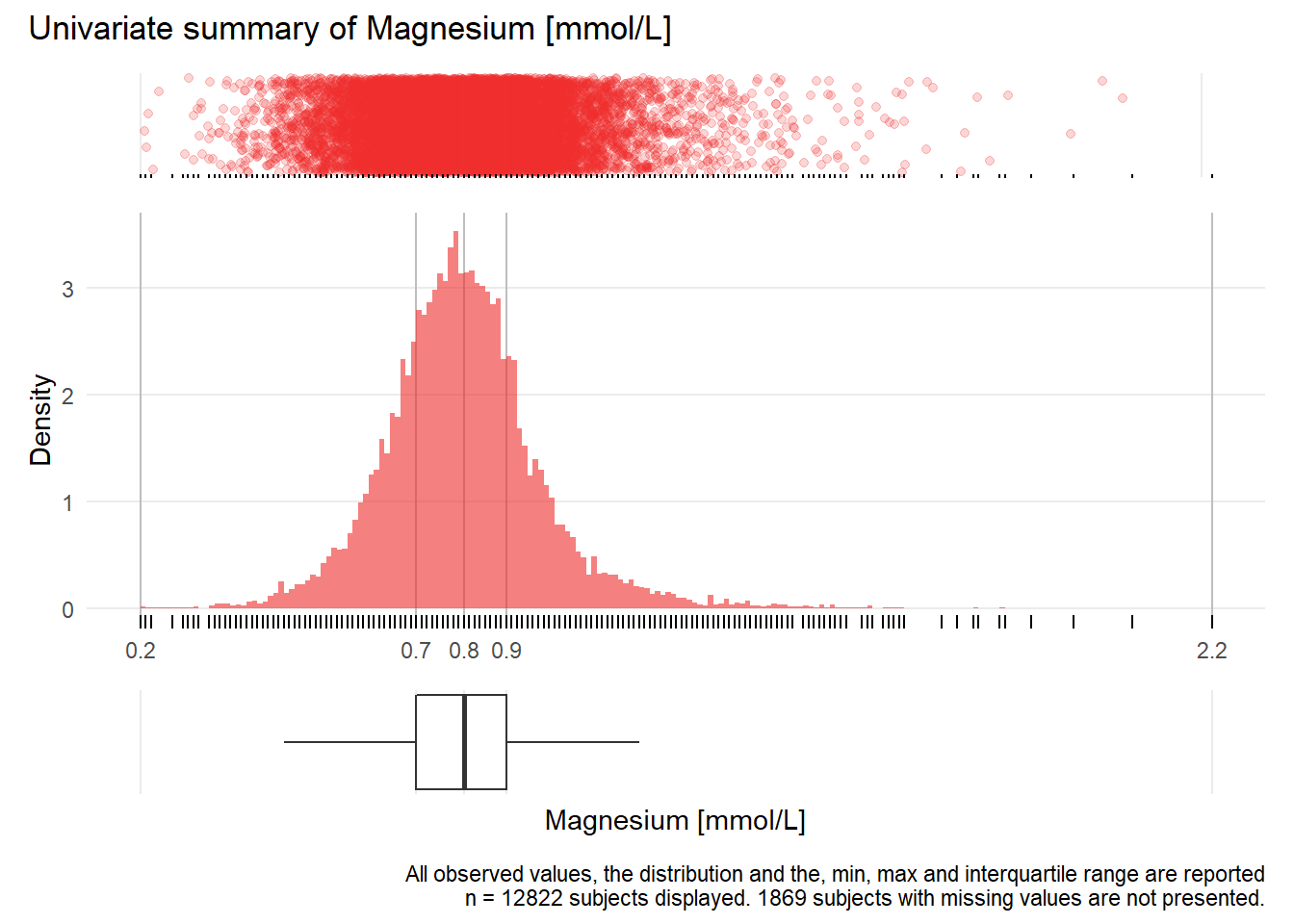
## Warning: Removed 2 rows containing missing values (geom_point).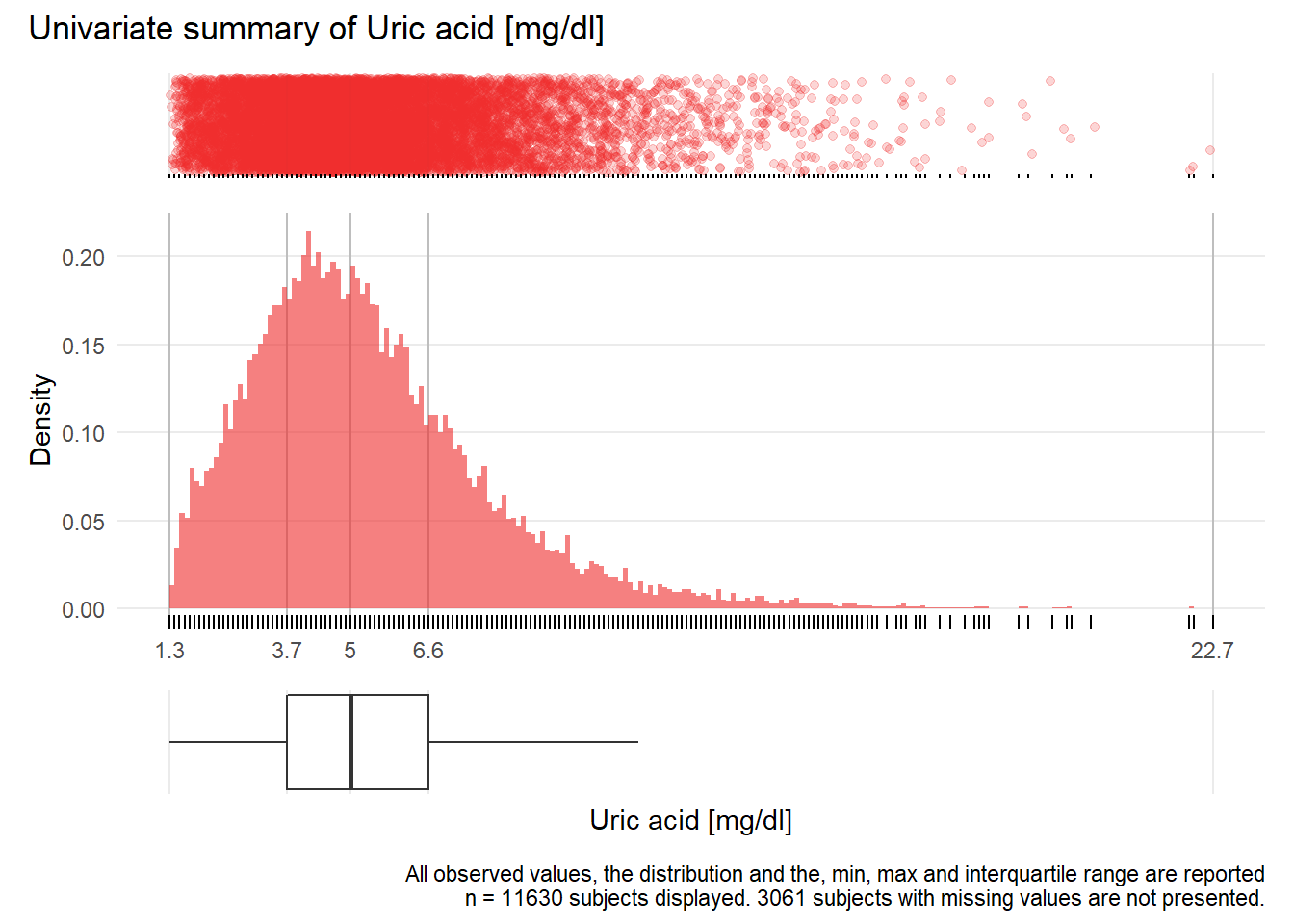
## Warning: Removed 4 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).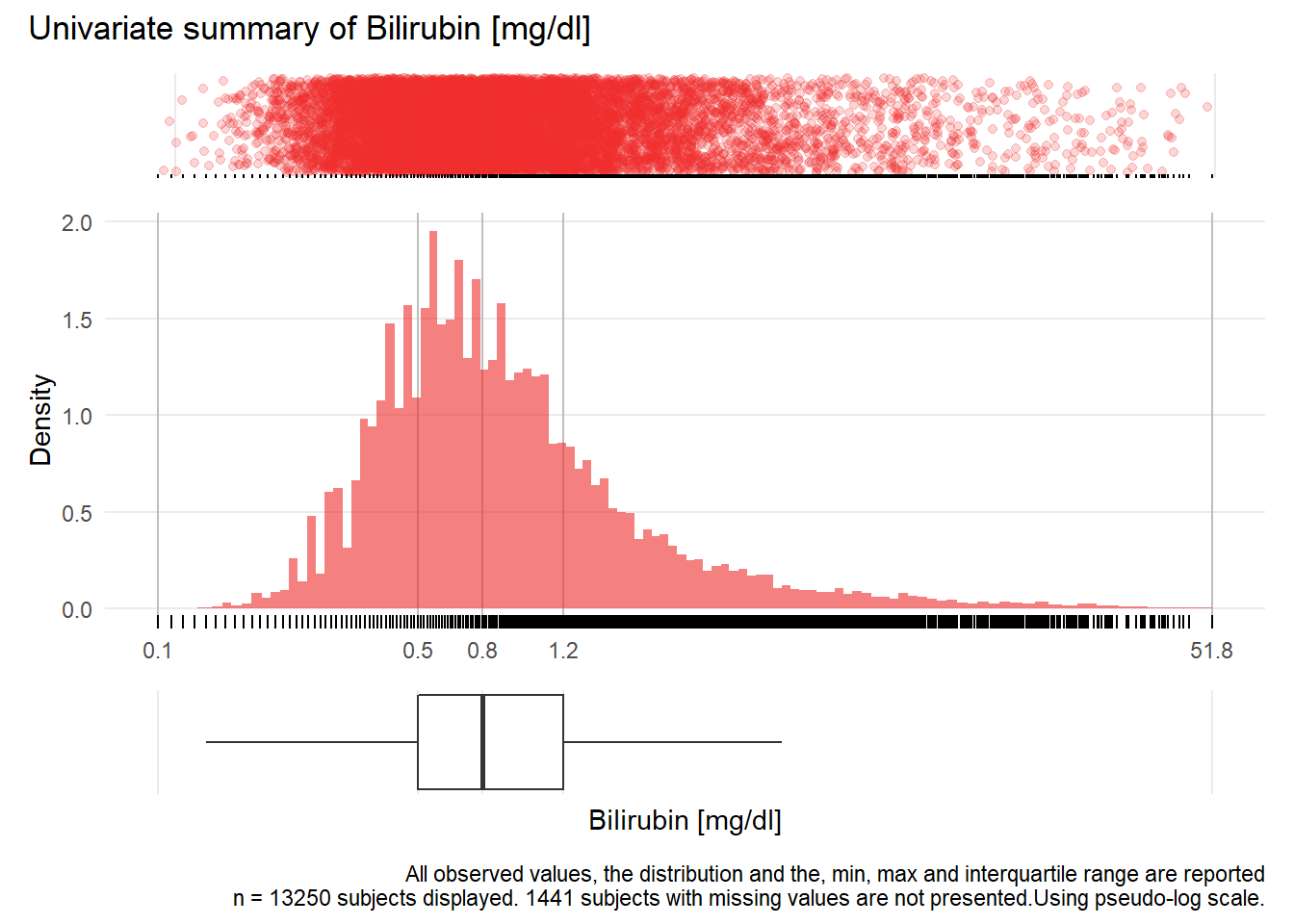
## Warning: Removed 2 rows containing missing values (geom_point).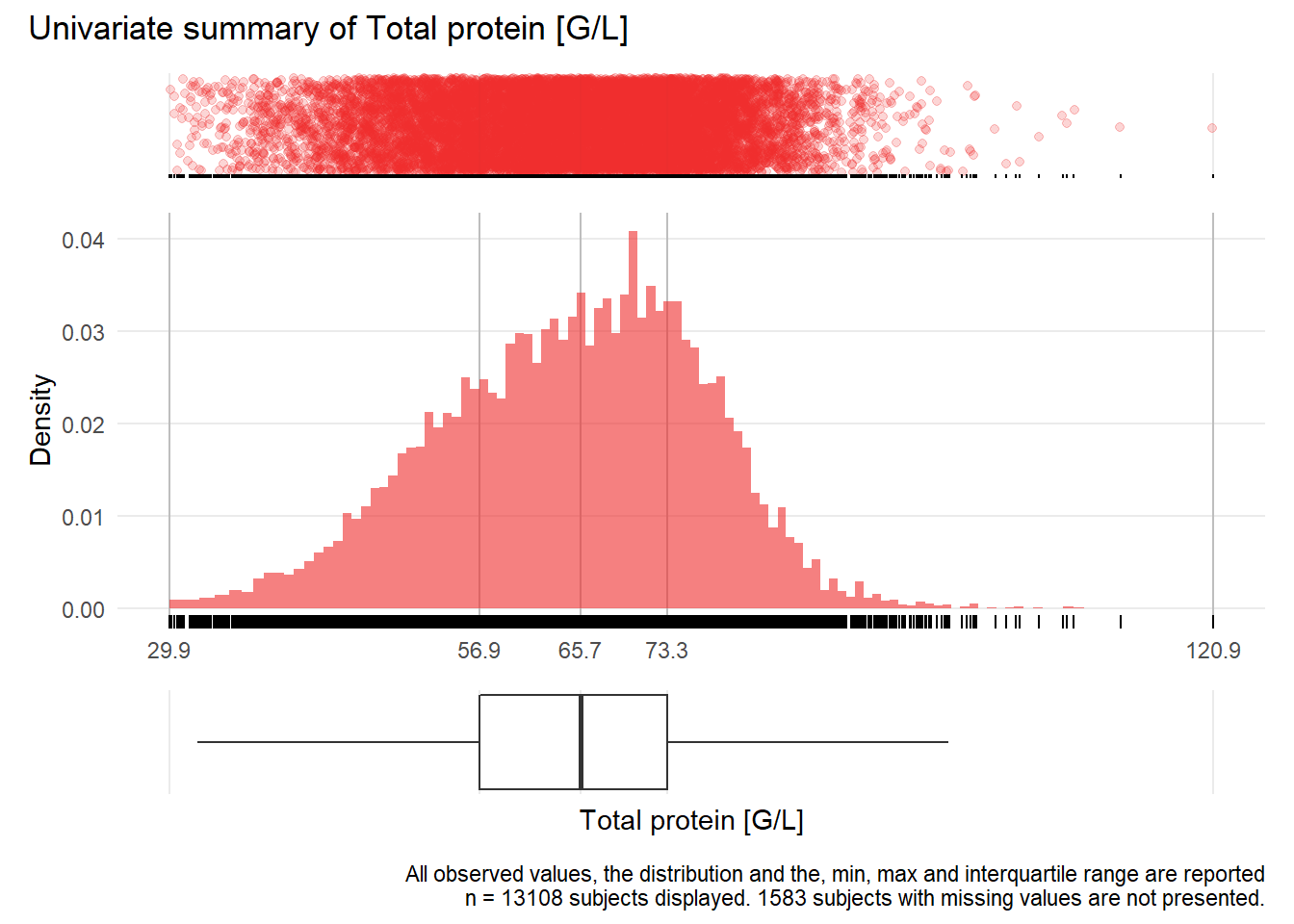
## Warning: Removed 1 rows containing missing values (geom_point).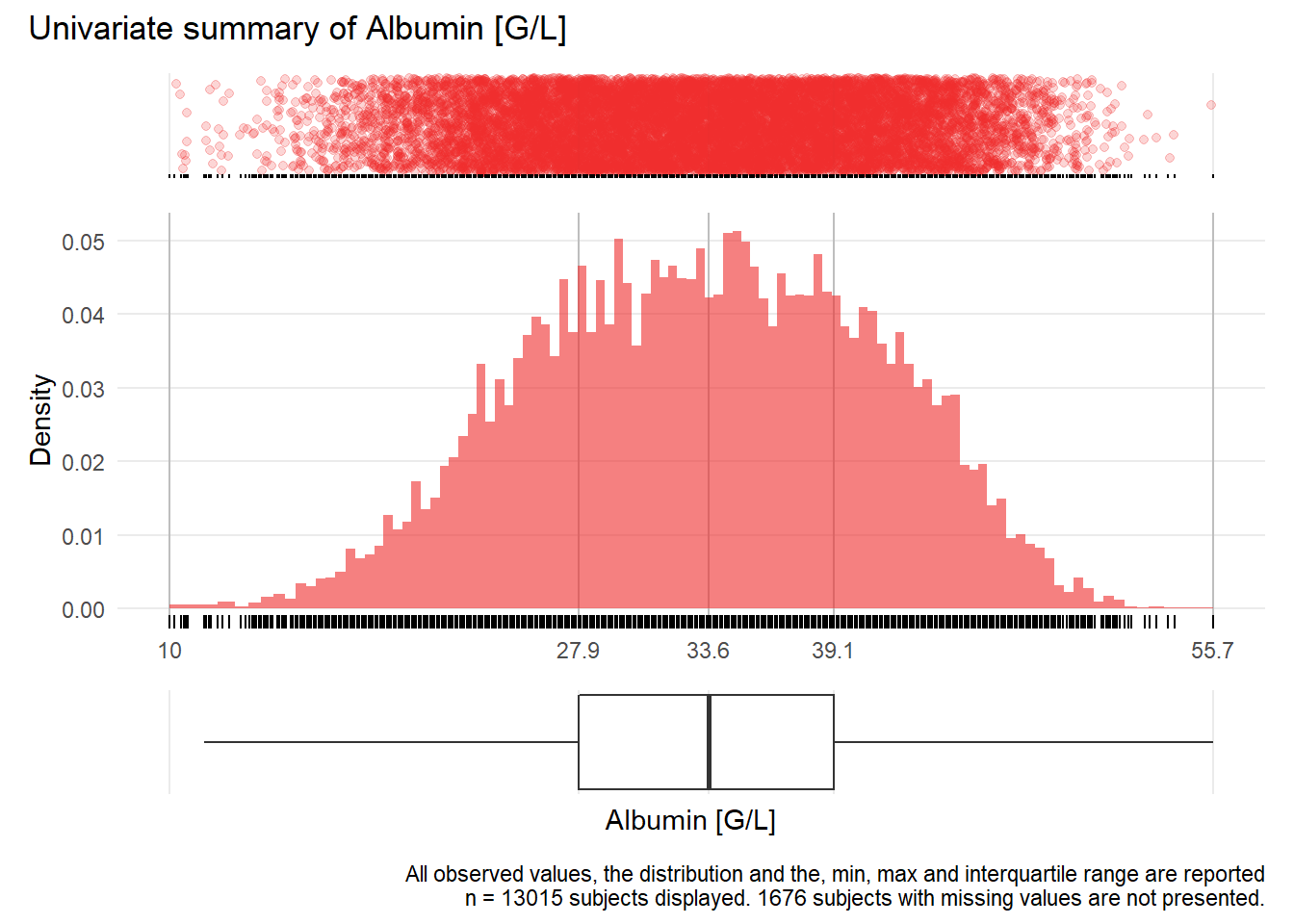
## Warning: Removed 9 rows containing missing values (geom_point).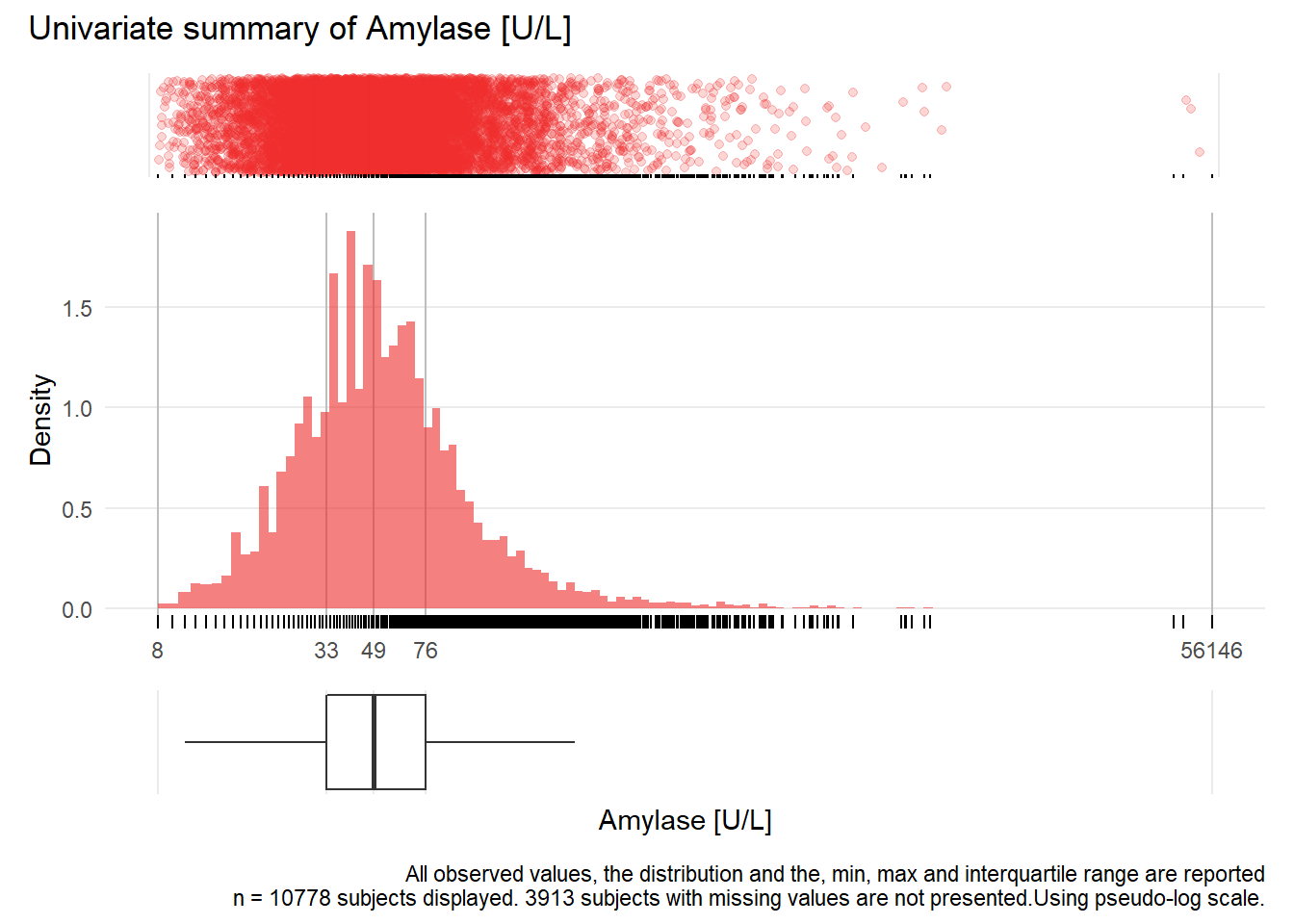
## Warning: Removed 3 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).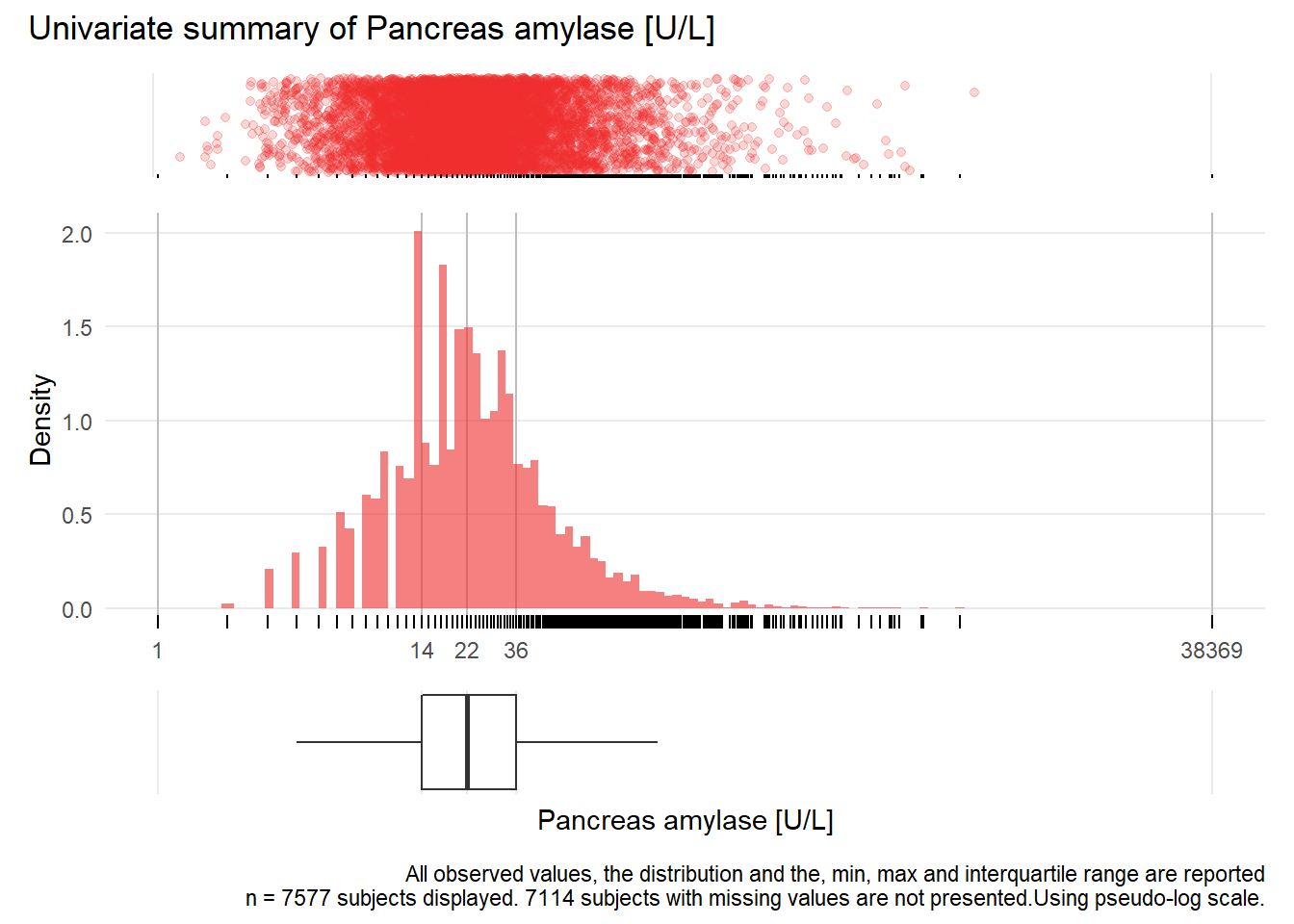
## Warning: Removed 2 rows containing missing values (geom_point).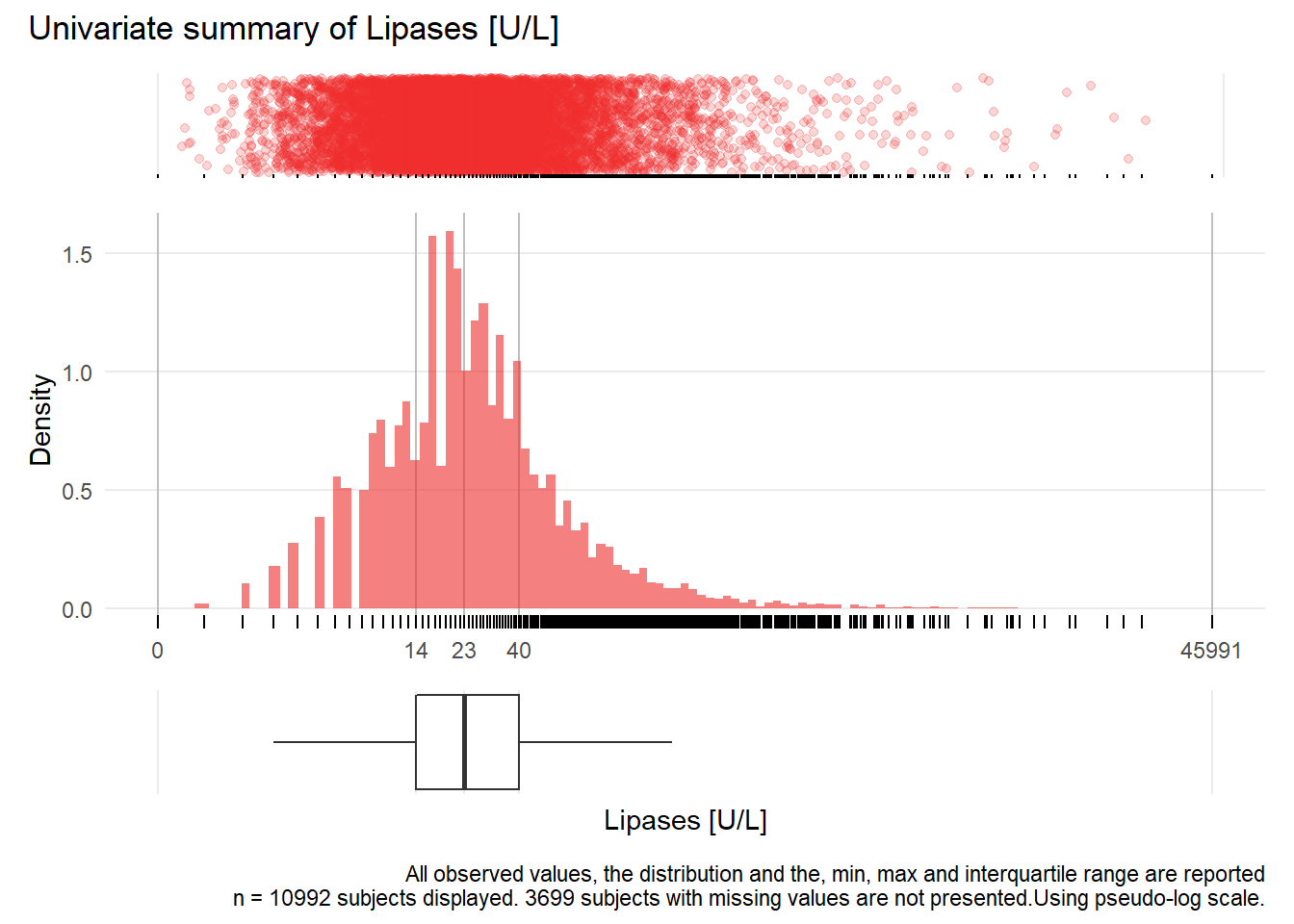
## Warning: Removed 17 rows containing missing values (geom_point).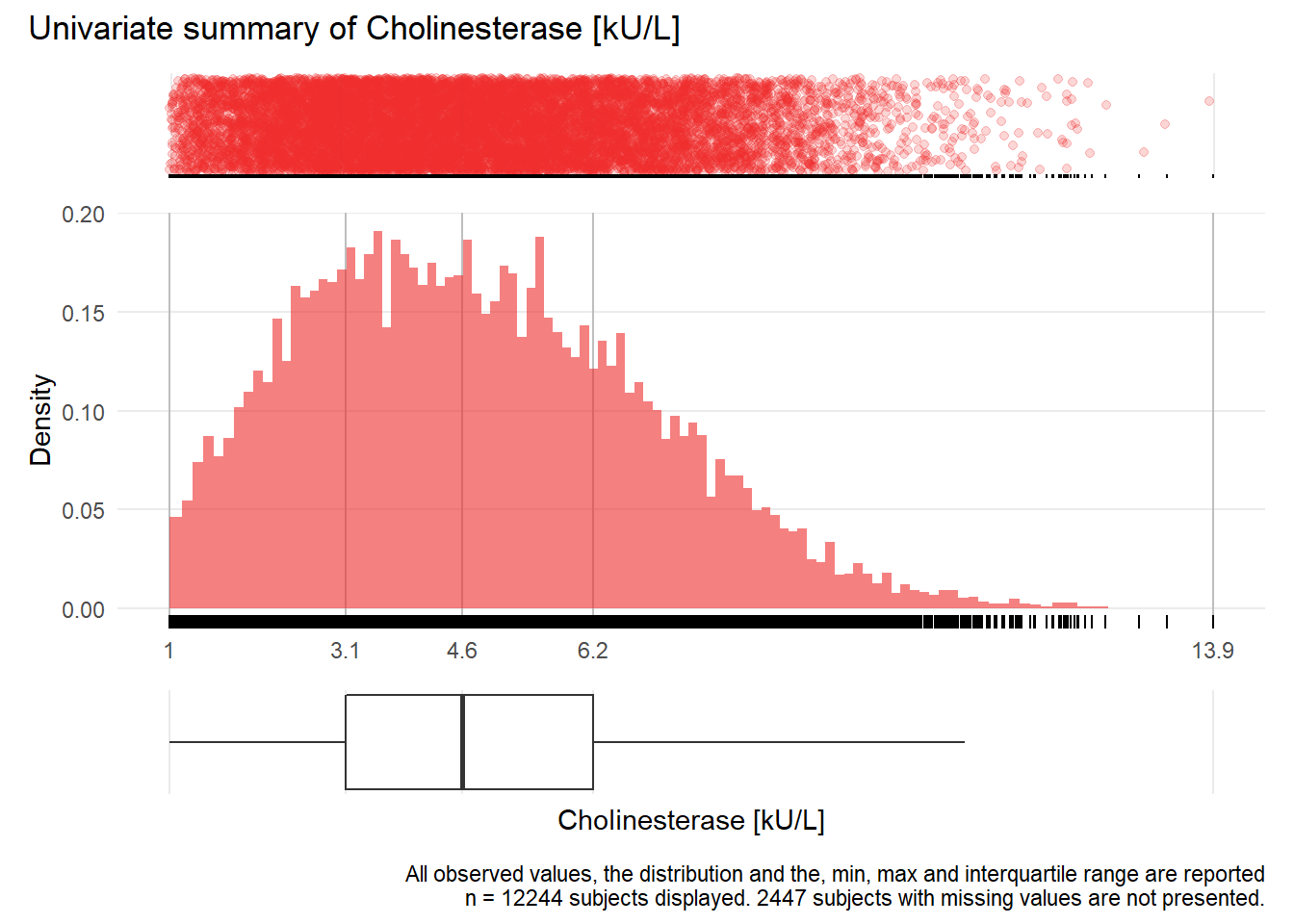
## Warning: Removed 2 rows containing missing values (geom_point).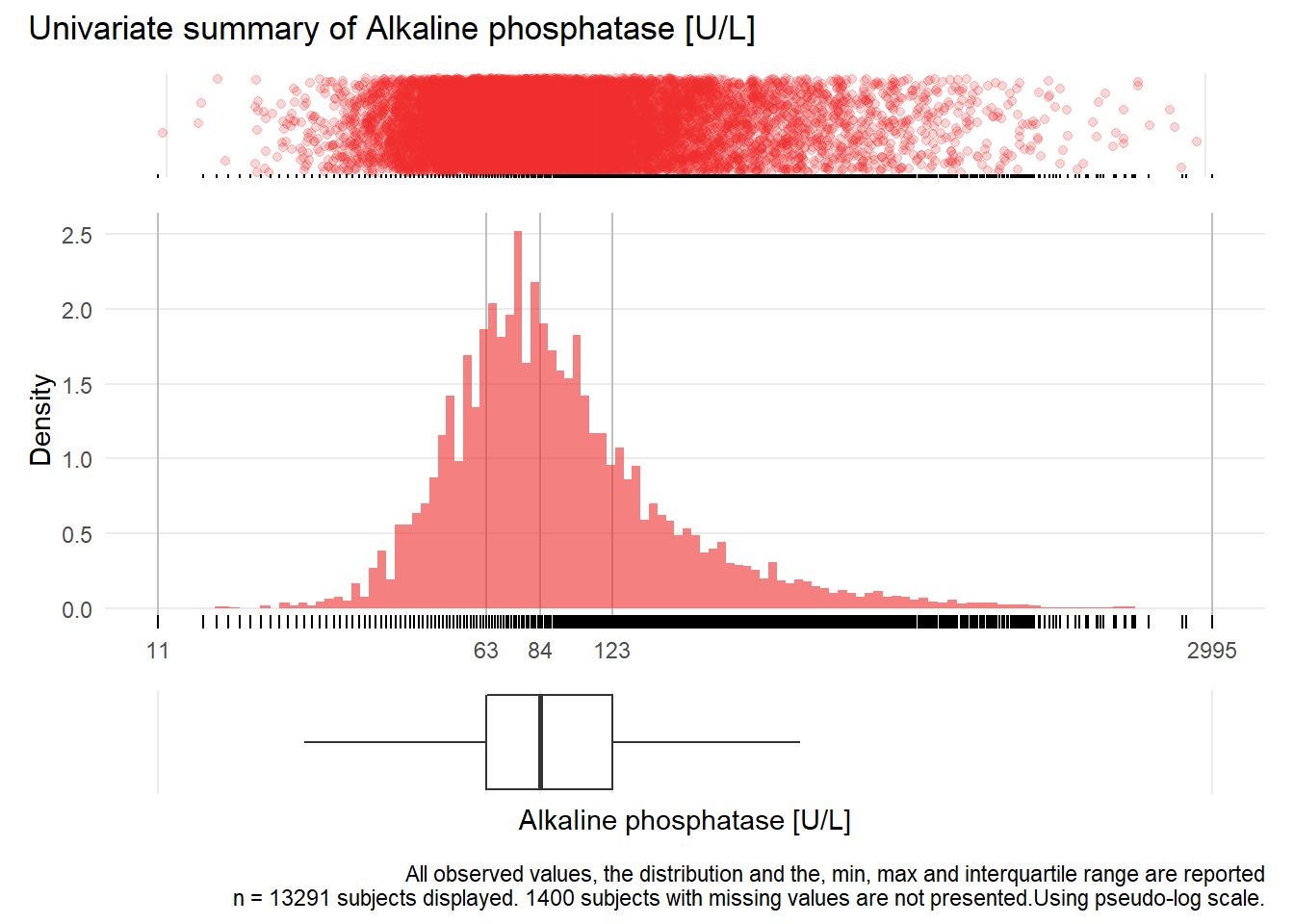
## Warning: Removed 1 rows containing missing values (geom_bar).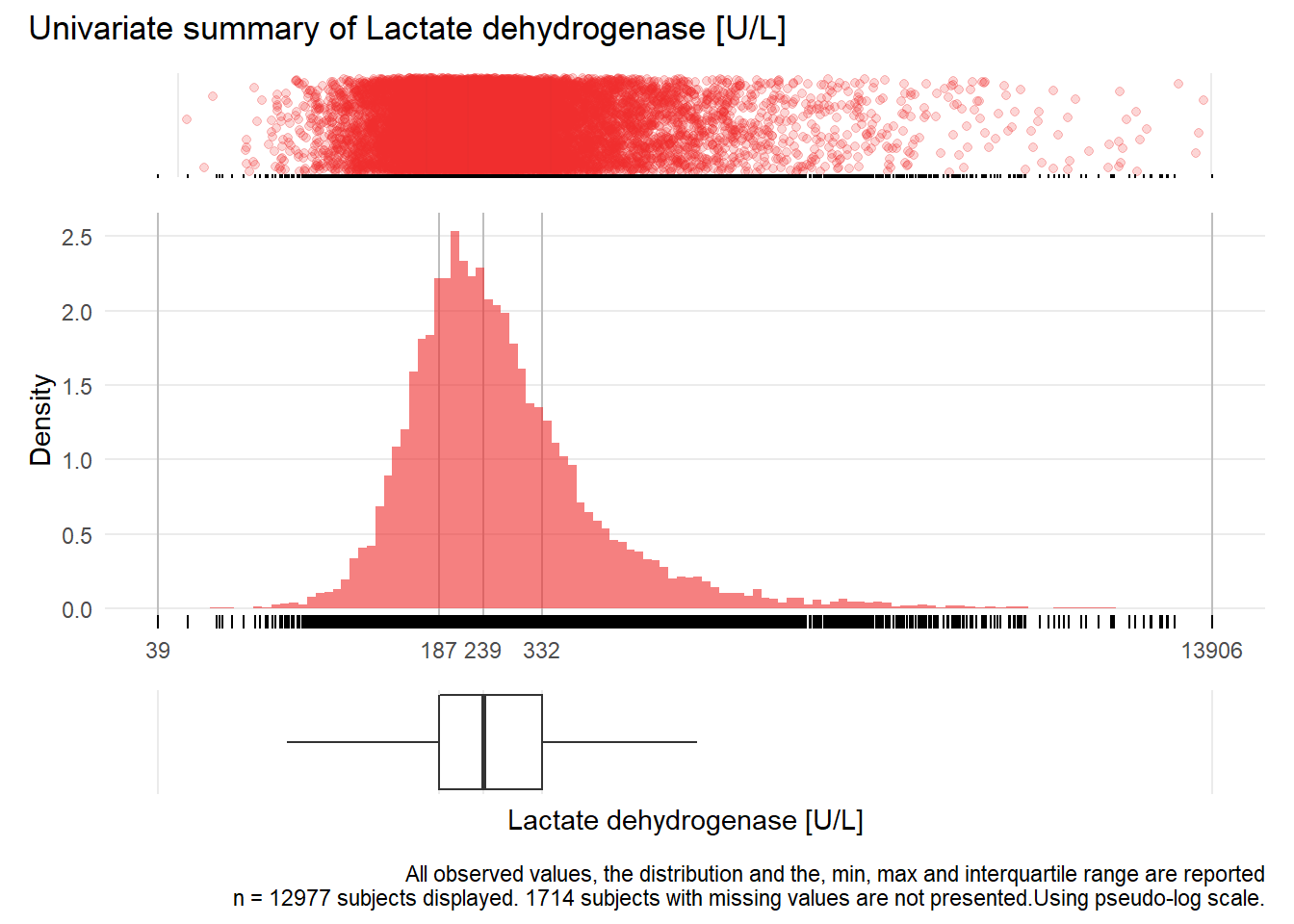
## Warning: Removed 7 rows containing missing values (geom_point).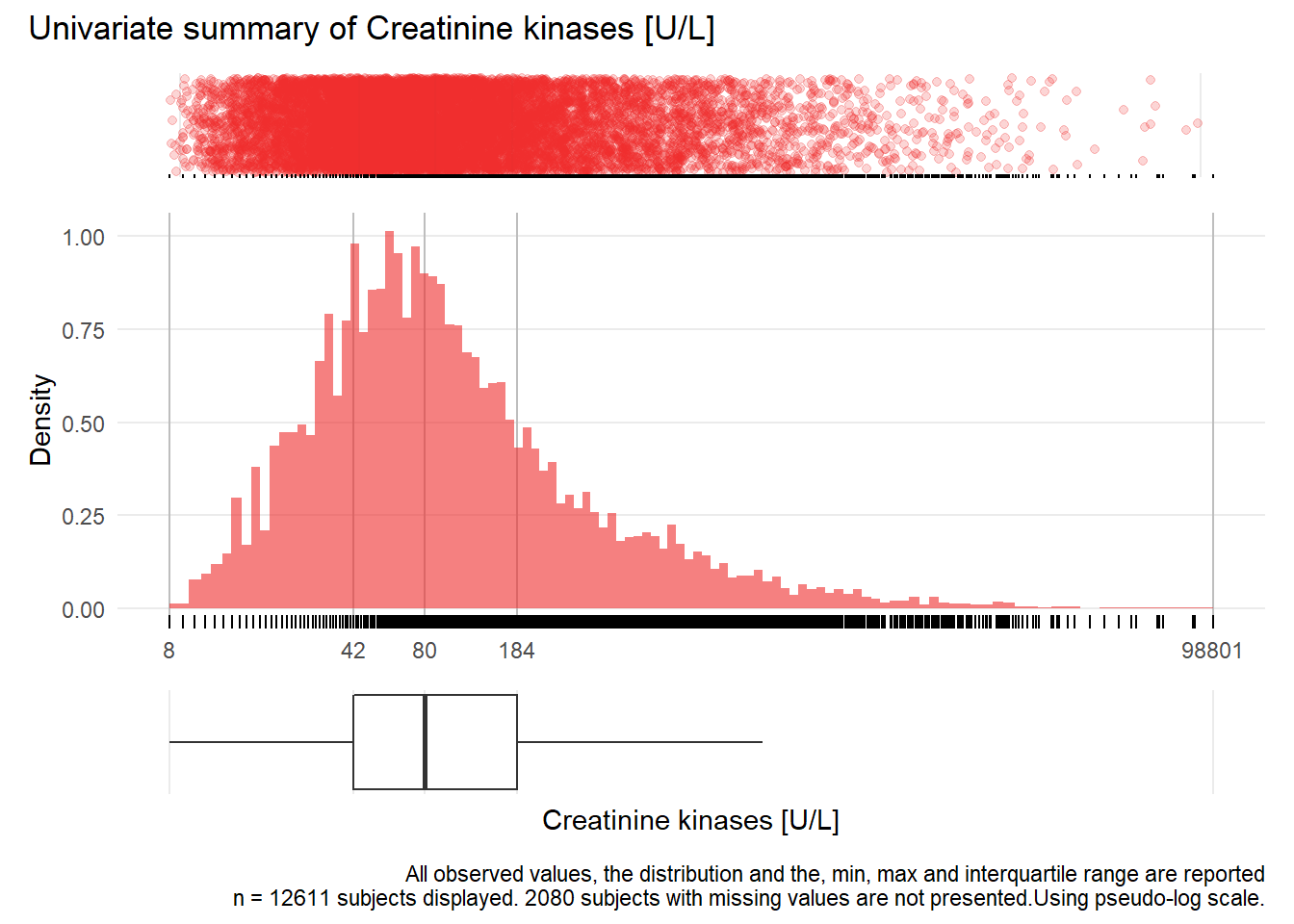
## Warning: Removed 2 rows containing missing values (geom_point).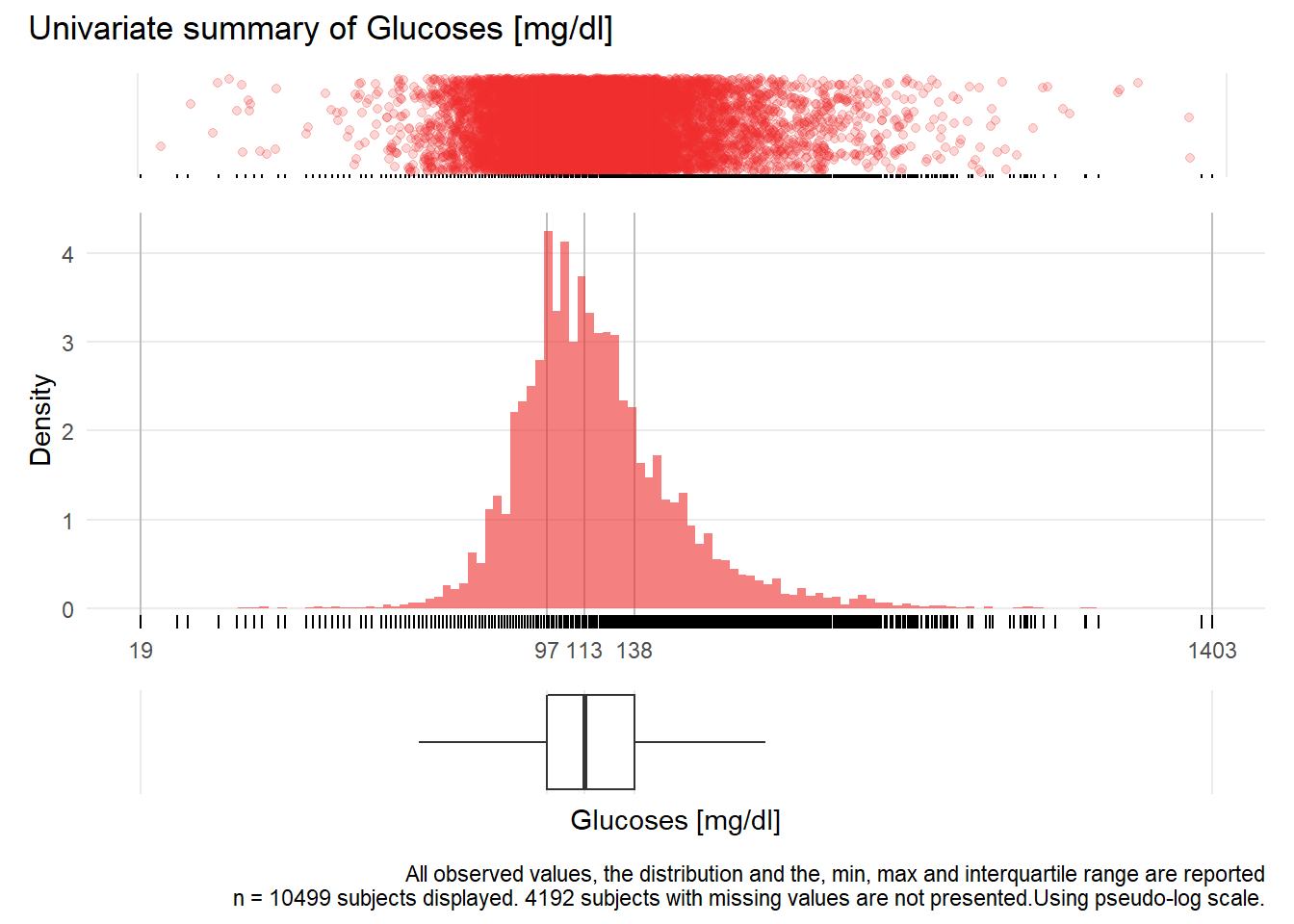
## Warning: Removed 3 rows containing missing values (geom_point).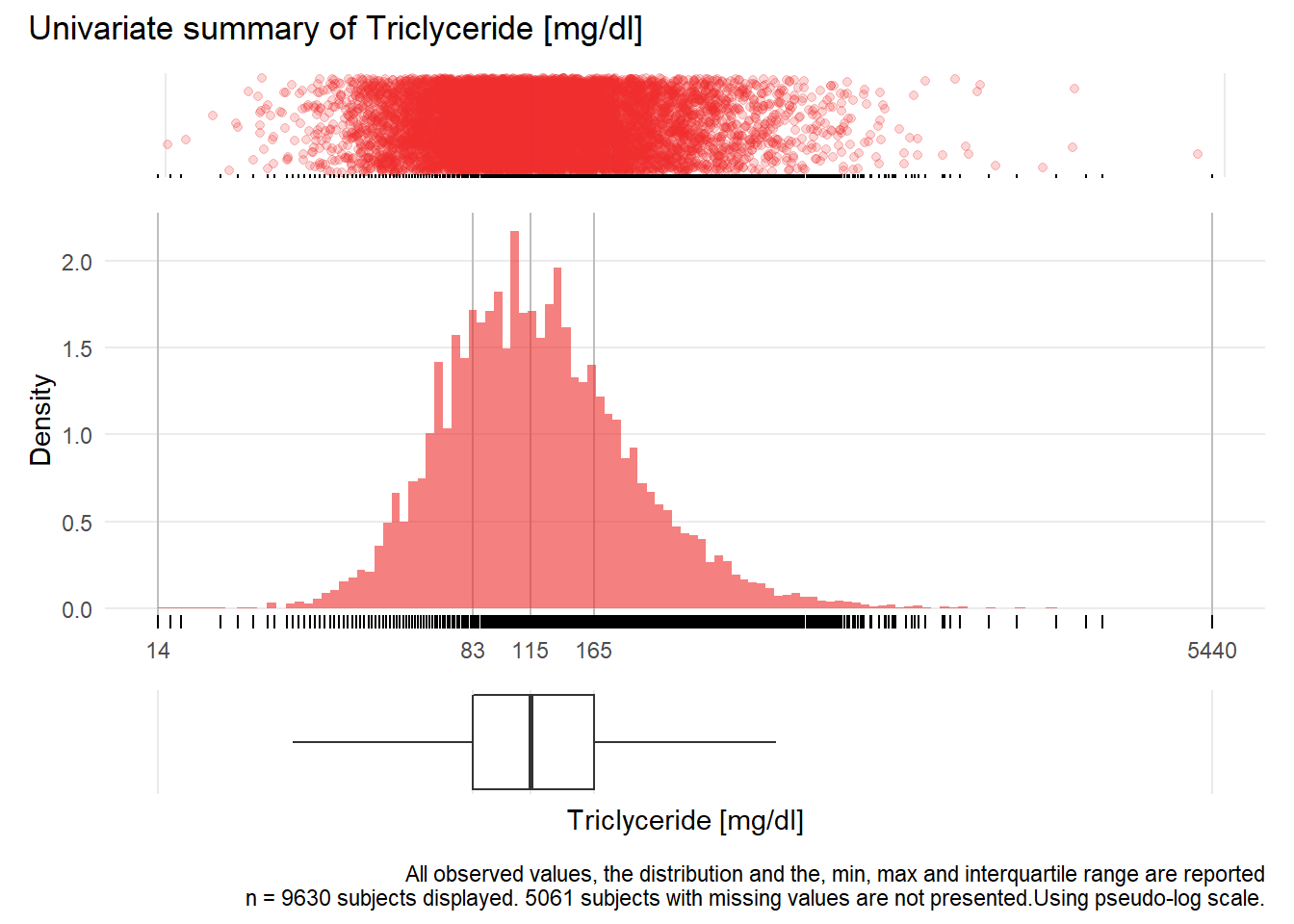
## Warning: Removed 1 rows containing missing values (geom_point).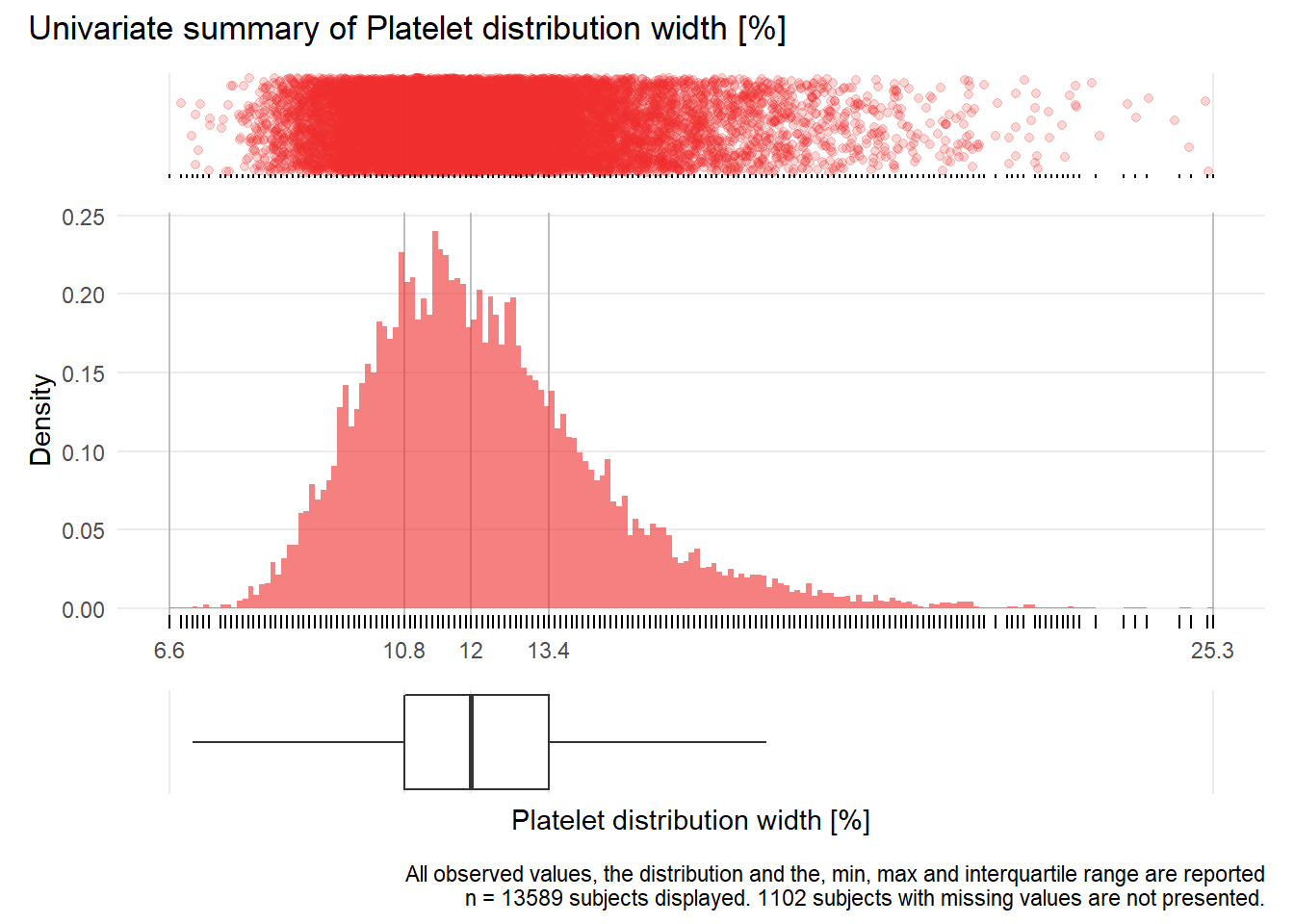
## Warning: Removed 2 rows containing missing values (geom_point).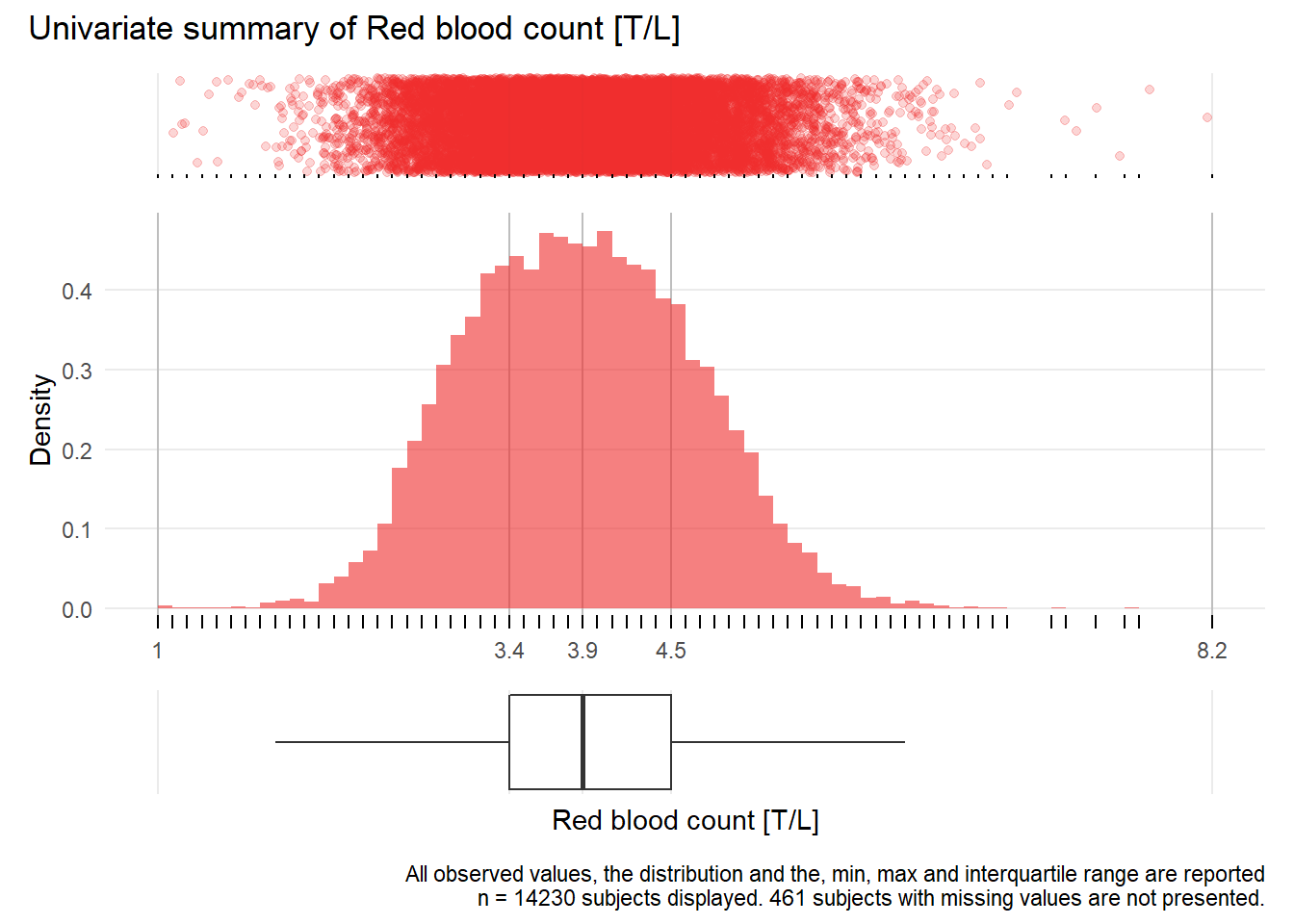
save(list=c("c_bact", "bact_variables", "sigma_values", "bact_transformed"),
file=here::here("data", "bact_env_c.rda"))17.3.3 Univariate distribution with variables using only the original variable without the suggested transformations
for(j in 1:length(unique.variables)){
print(ida_plot_univar(b_bact, unique.variables[j], sigma=res[j], n_bars=100, transform = FALSE))
# if(!is.na(res[j])){
# print(ida_plot_univar(b_bact, paste("t_",variables[j],sep="")))
# }
}## Warning: Removed 3 rows containing missing values (geom_point).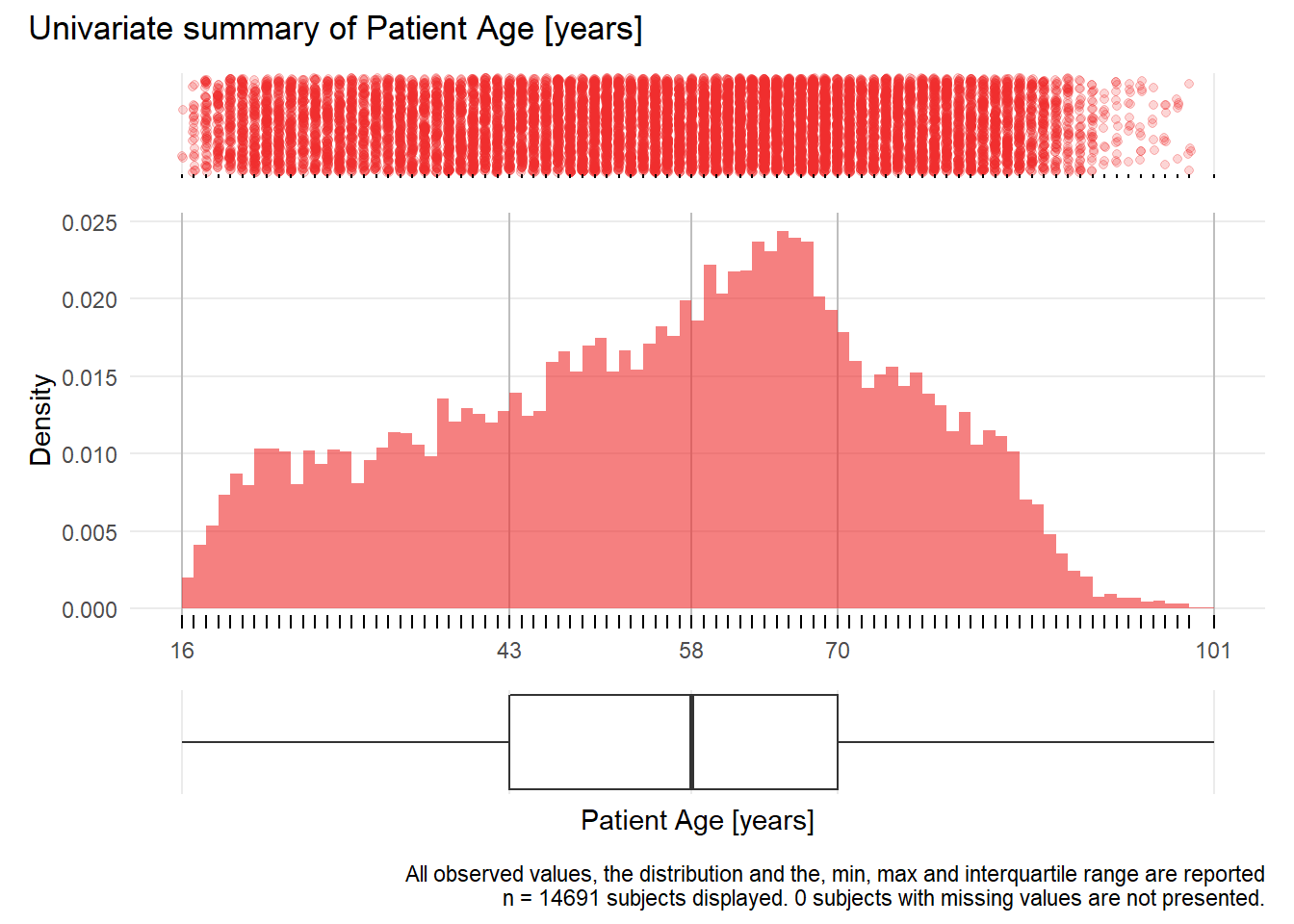
## Warning: Removed 6 rows containing missing values (geom_point).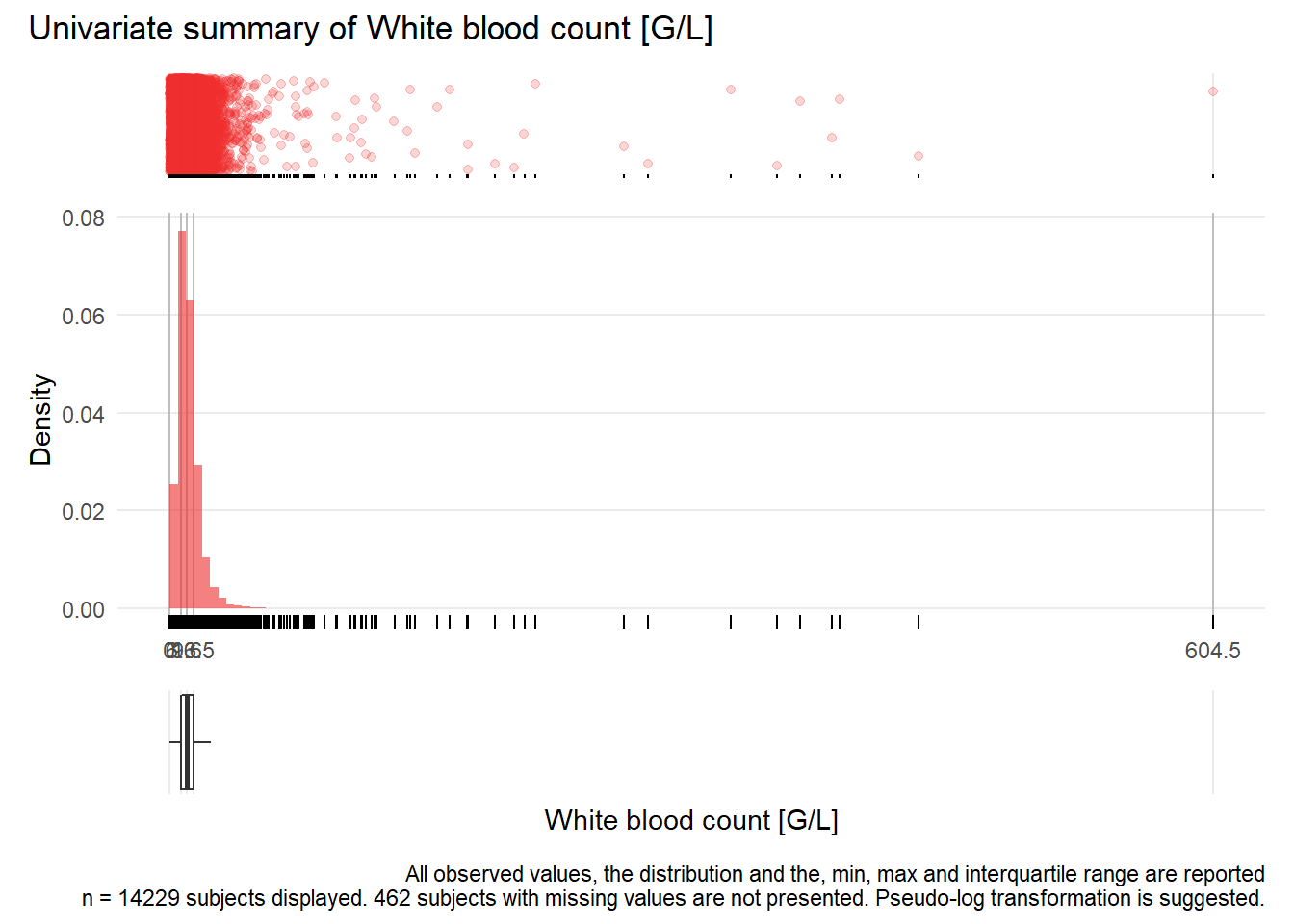
## Warning: Removed 1 rows containing missing values (geom_point).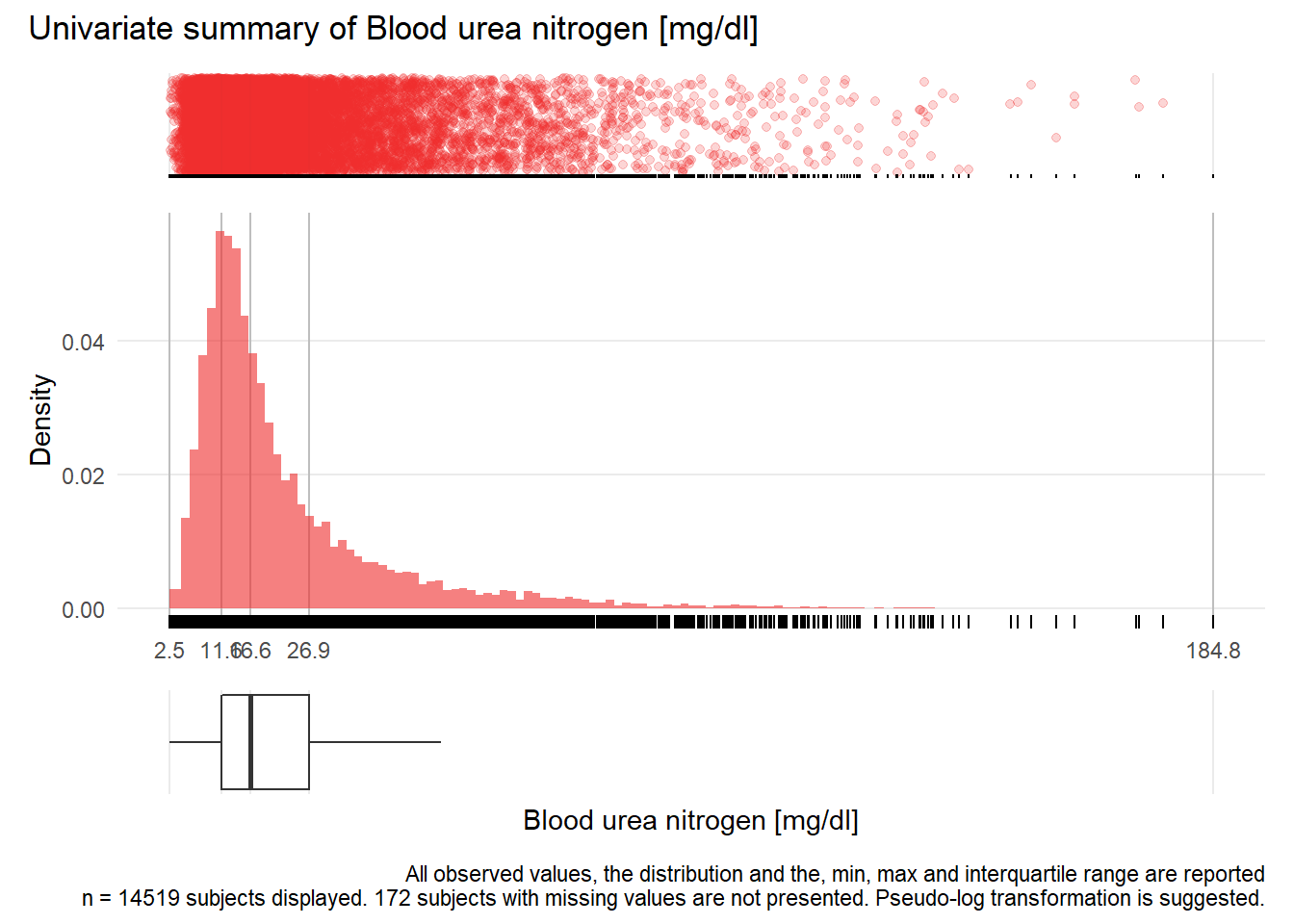
## Warning: Removed 3 rows containing missing values (geom_point).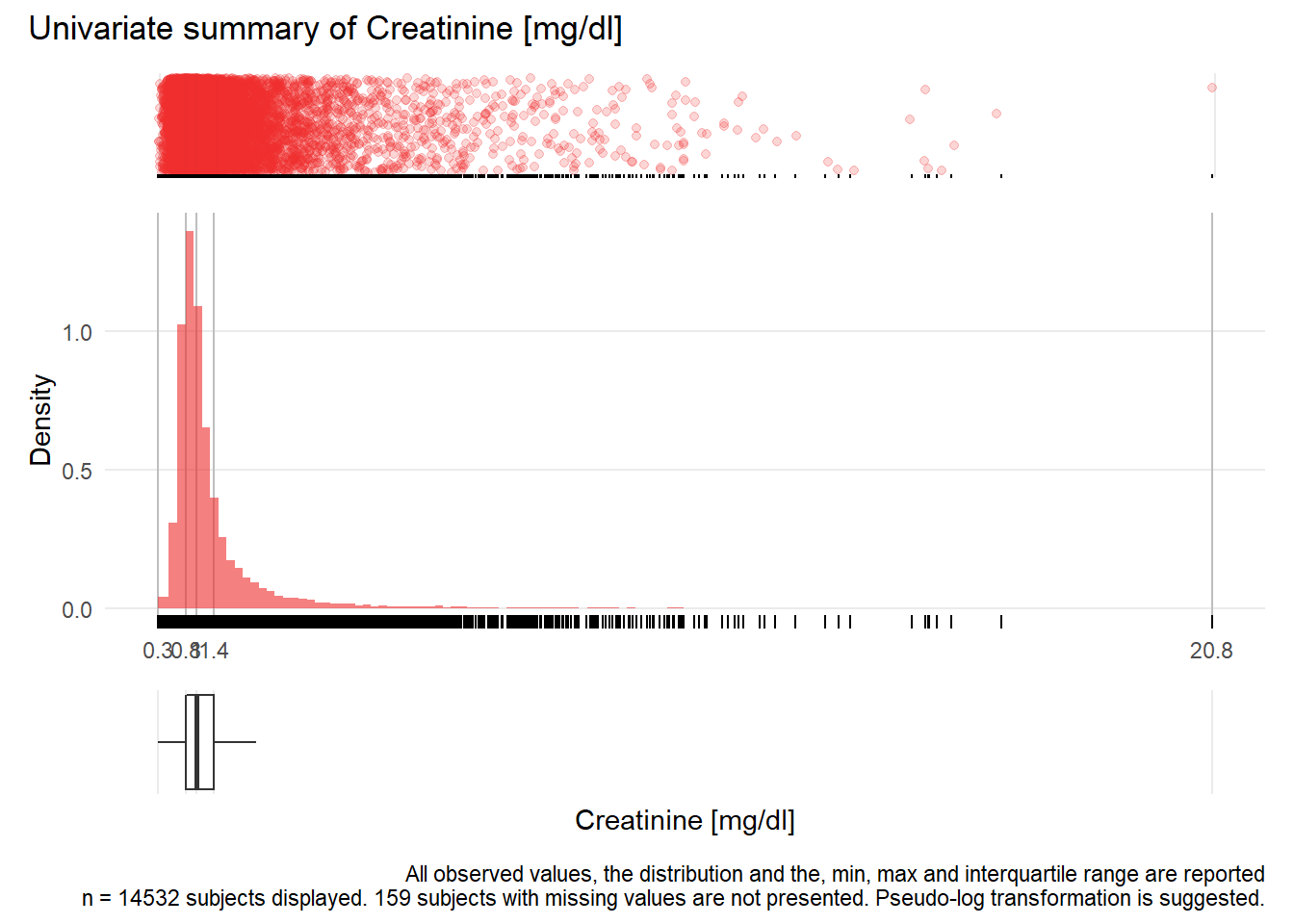
## Warning: Removed 59 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).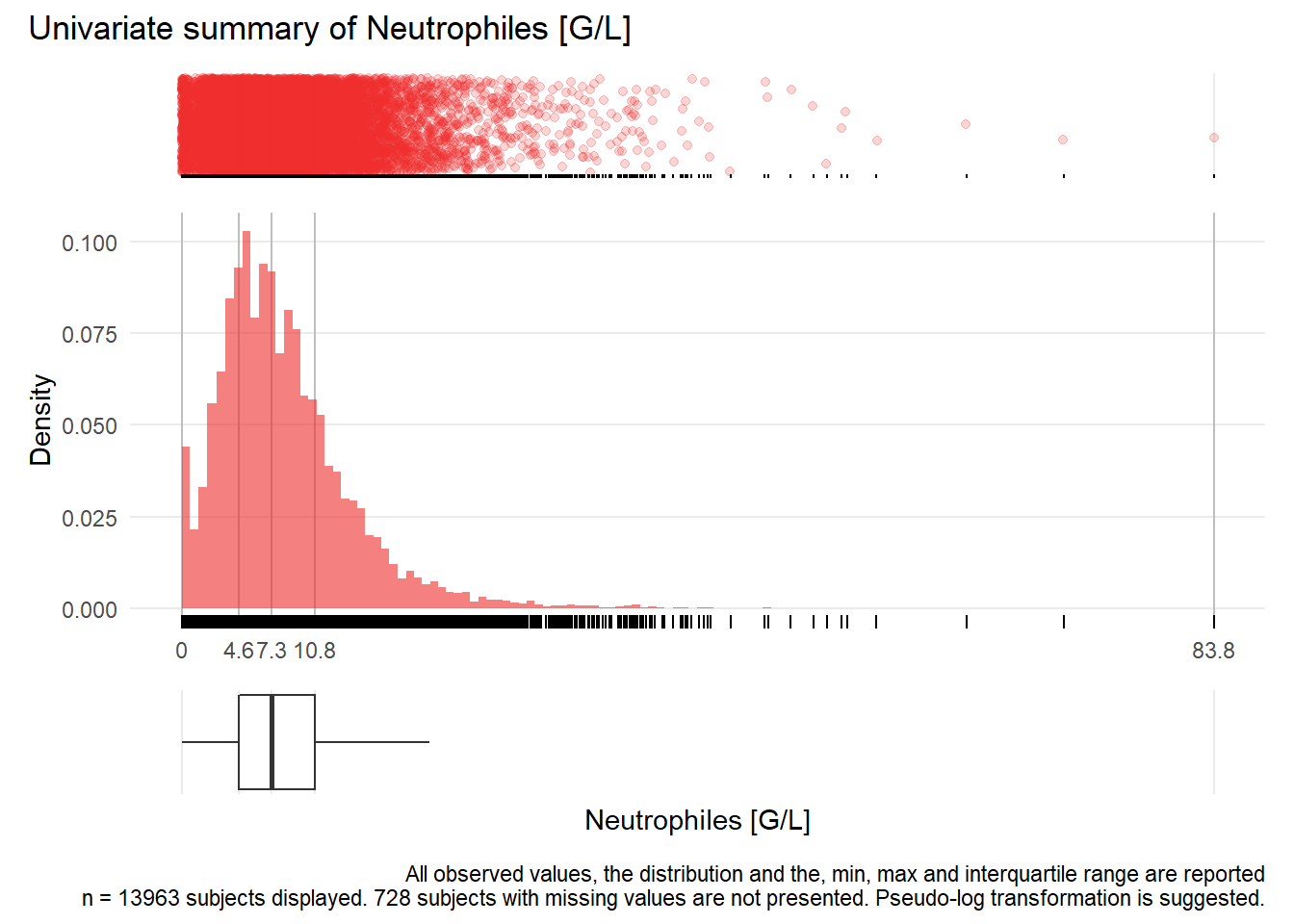
## Warning: Removed 3 rows containing missing values (geom_point).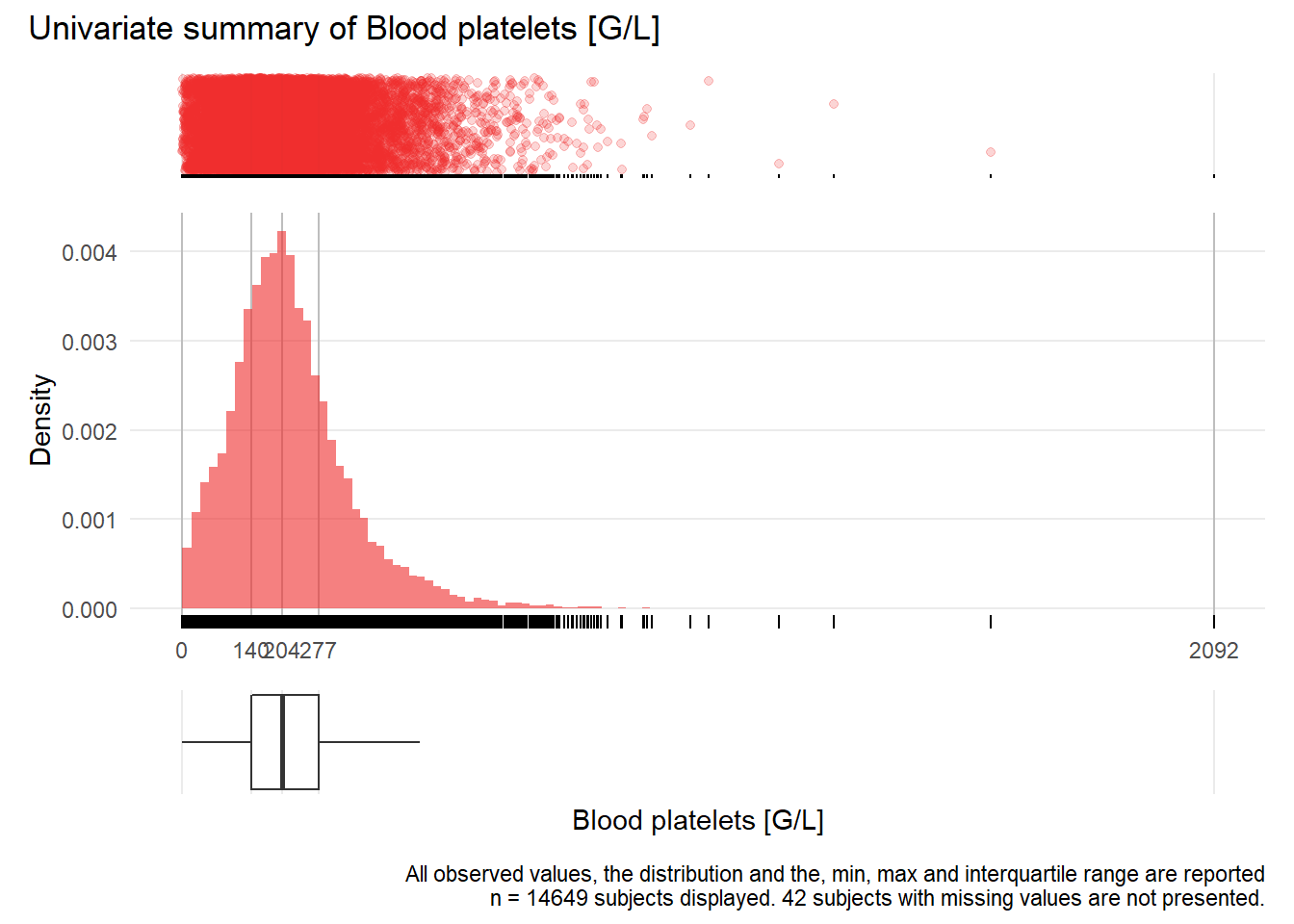
## Warning: Removed 3407 rows containing missing values (geom_point).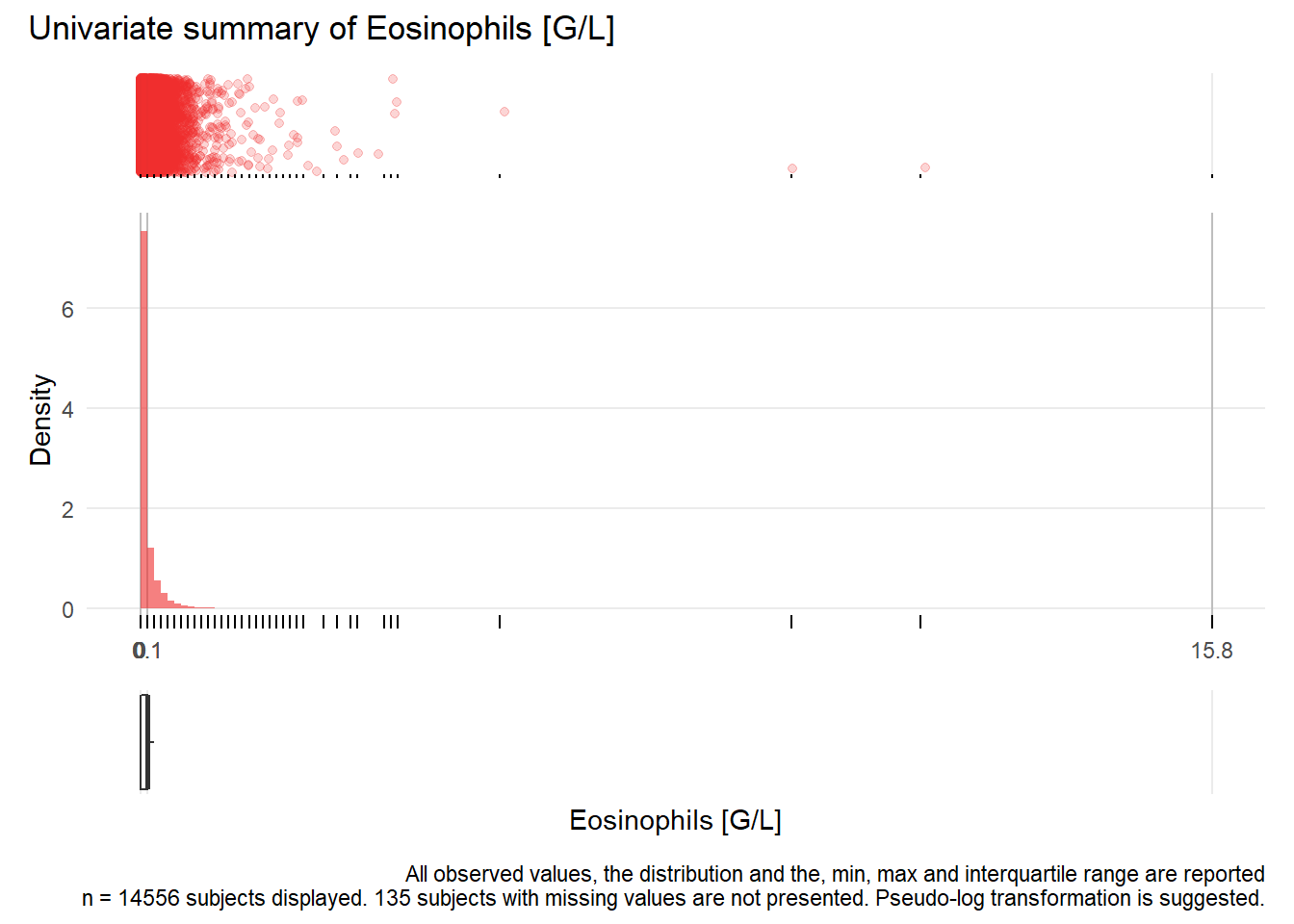
## Warning: Removed 6332 rows containing missing values (geom_point).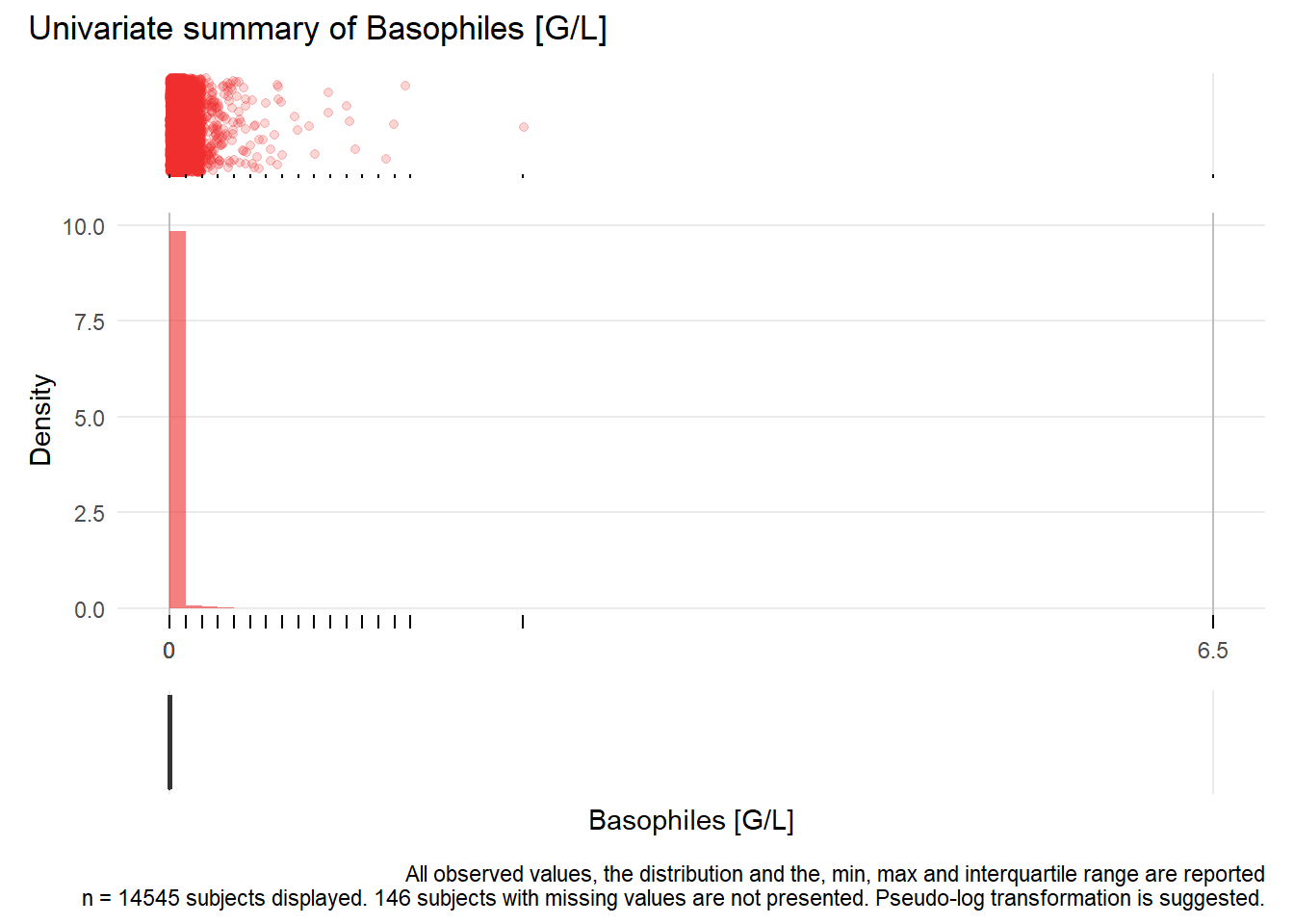
## Warning: Removed 54 rows containing missing values (geom_point).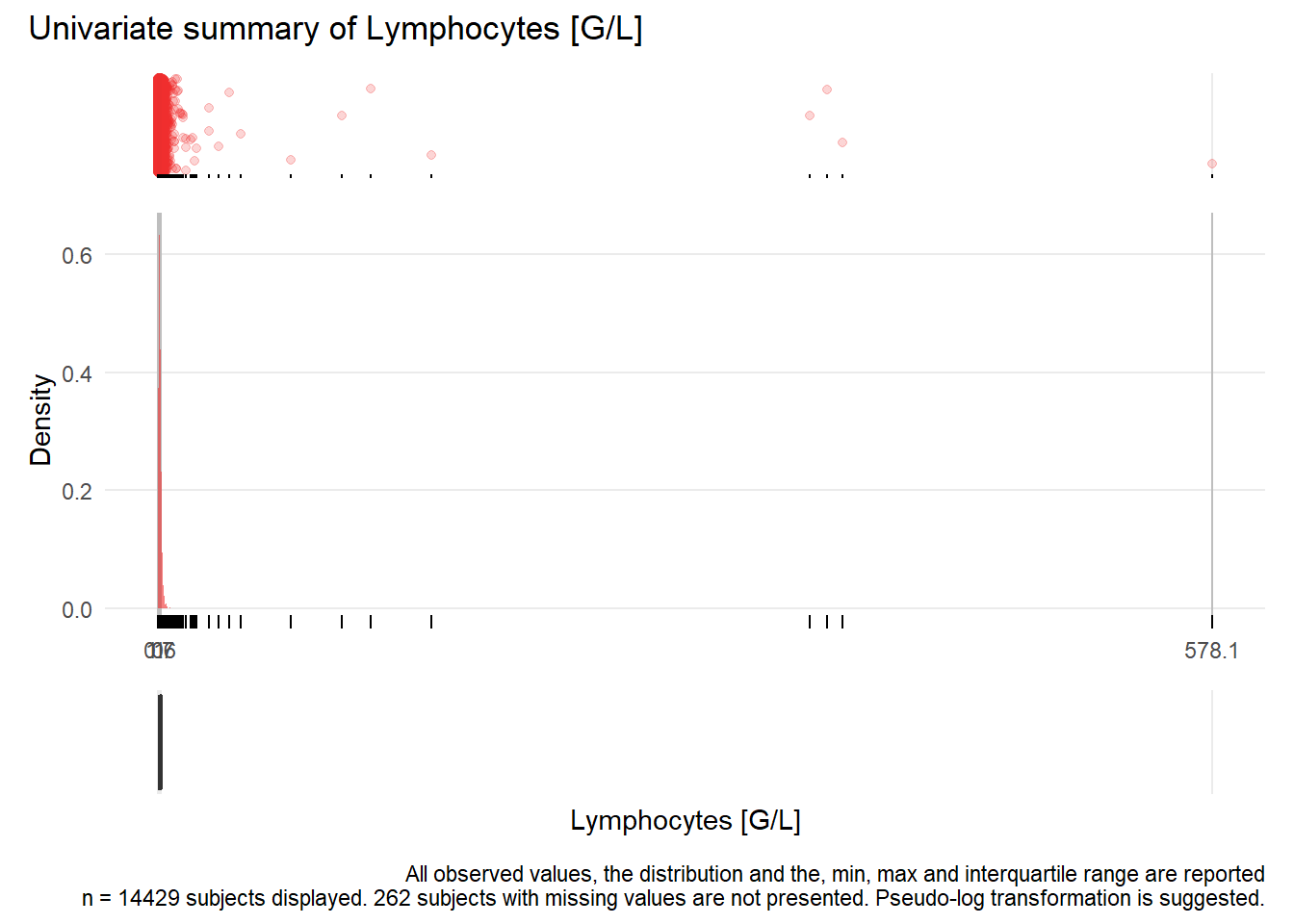
## Warning: Removed 188 rows containing missing values (geom_point).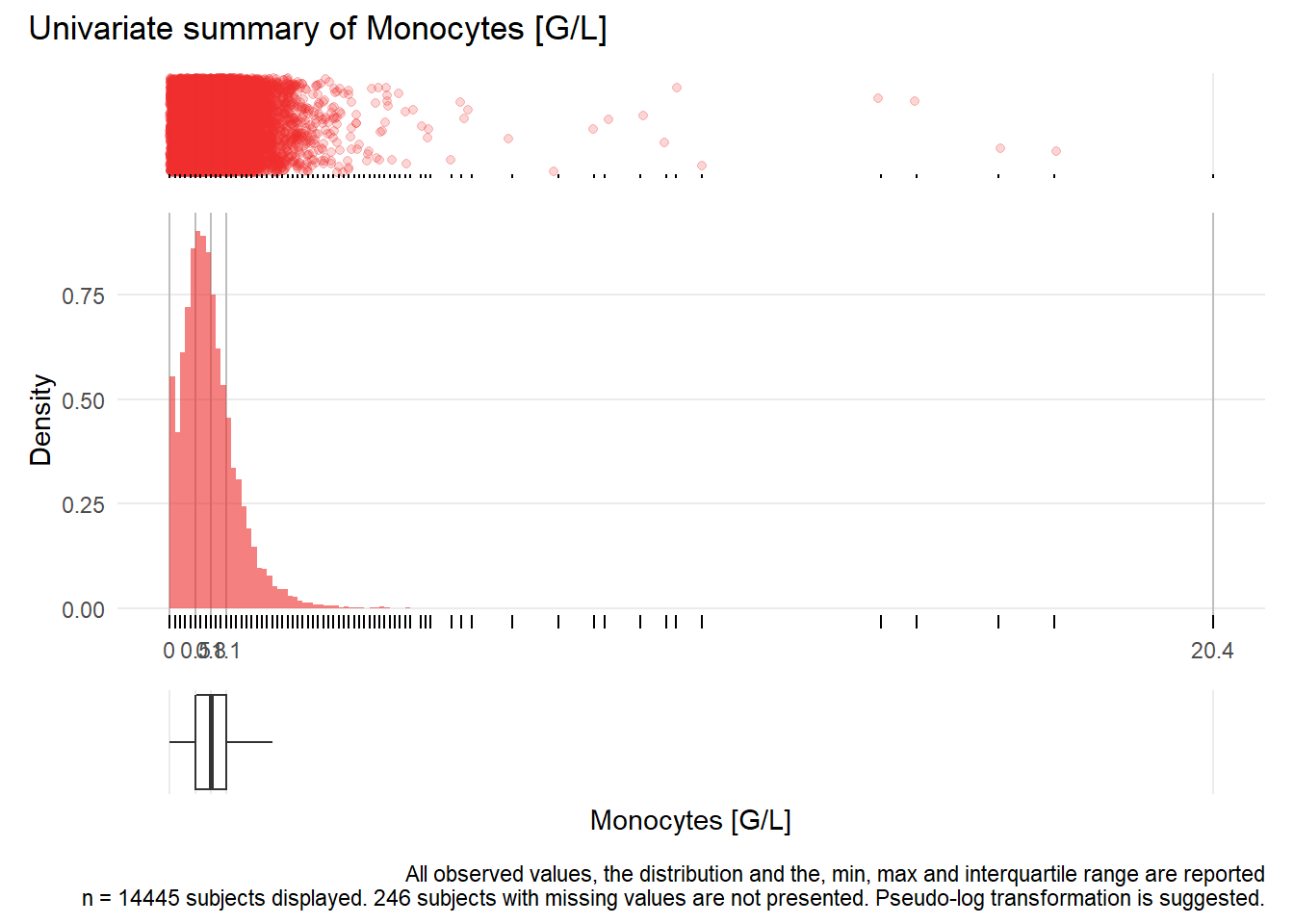
## Warning: Removed 82 rows containing missing values (geom_point).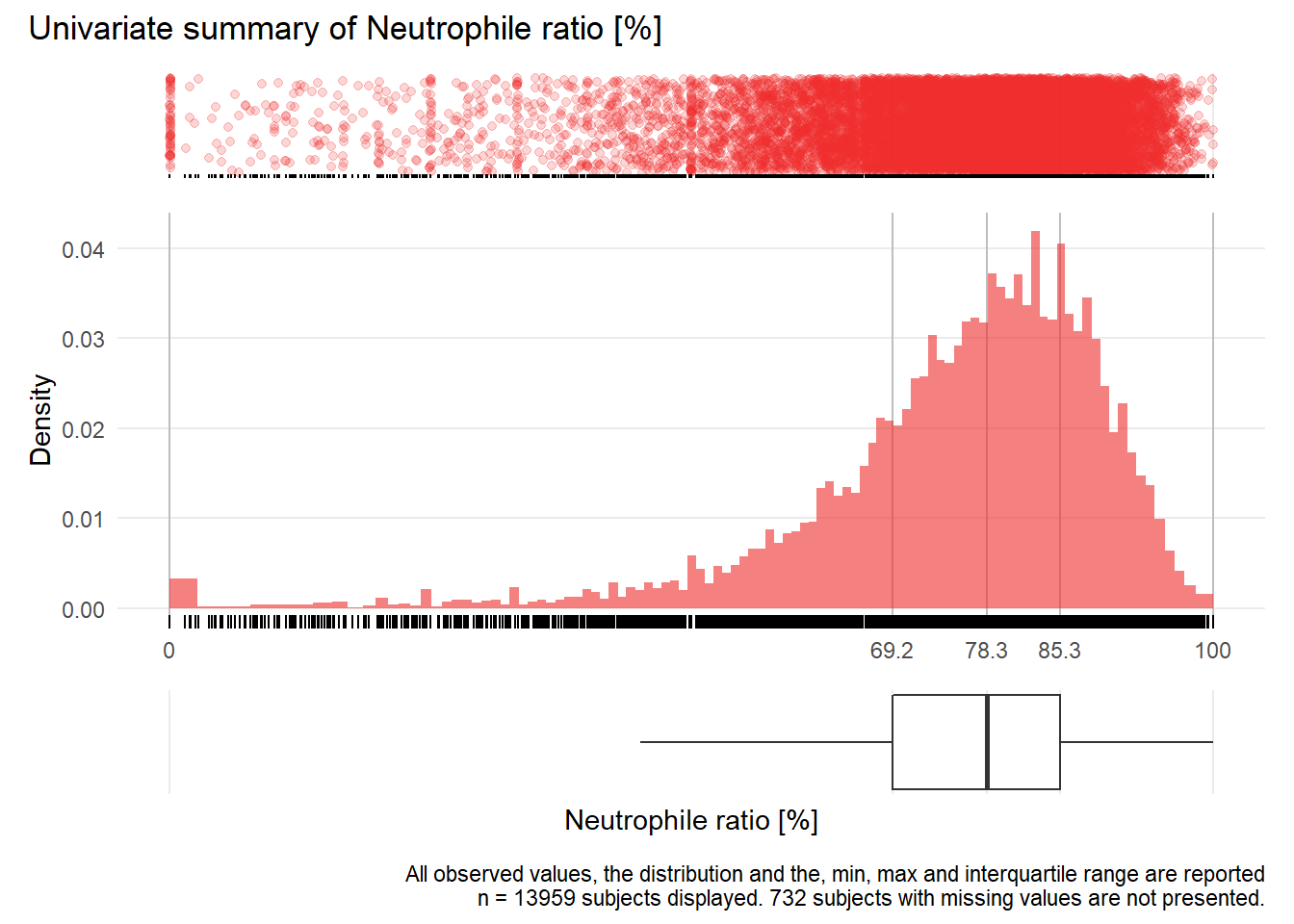
## Warning: Removed 3333 rows containing missing values (geom_point).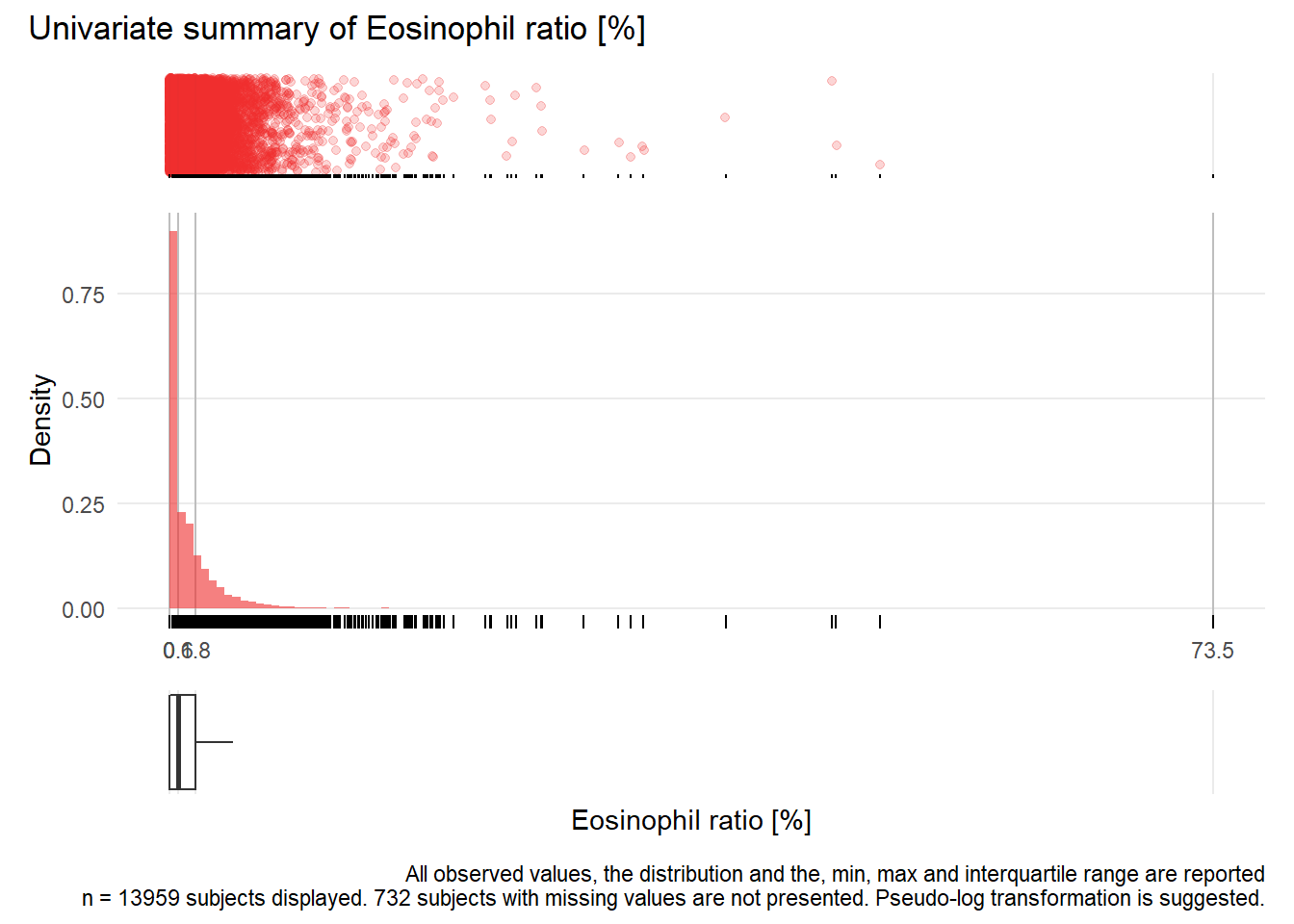
## Warning: Removed 6181 rows containing missing values (geom_point).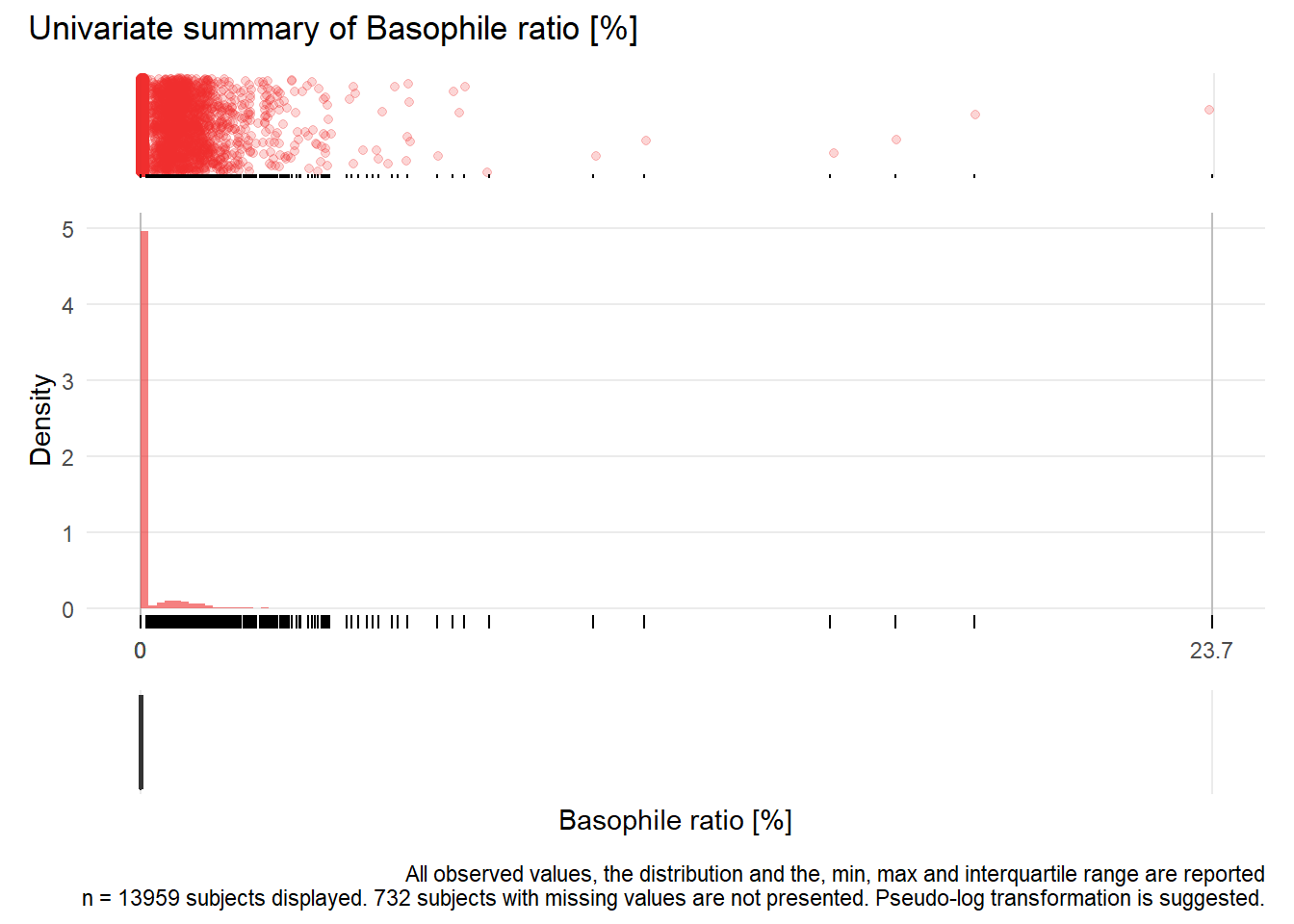
## Warning: Removed 68 rows containing missing values (geom_point).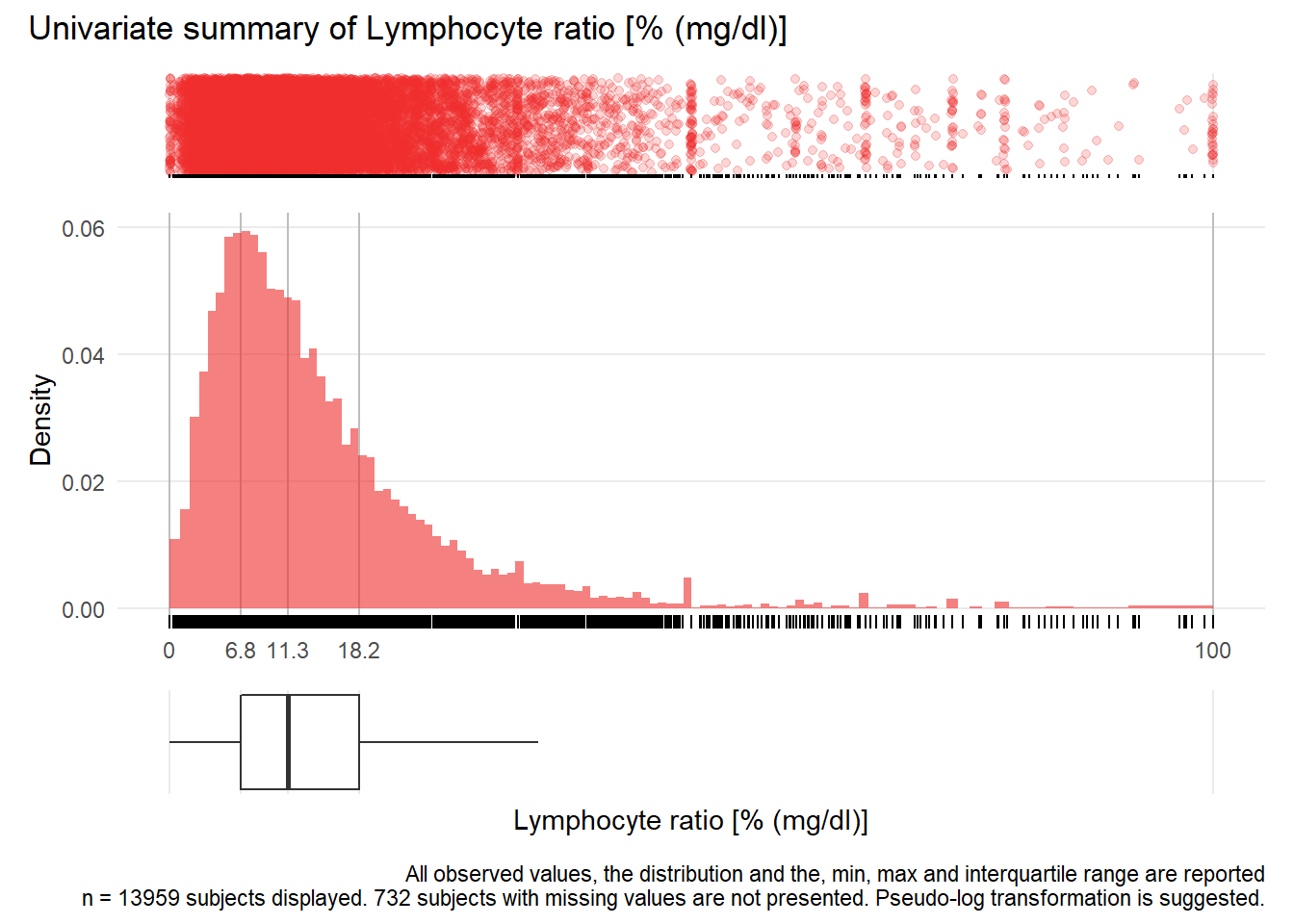
## Warning: Removed 177 rows containing missing values (geom_point).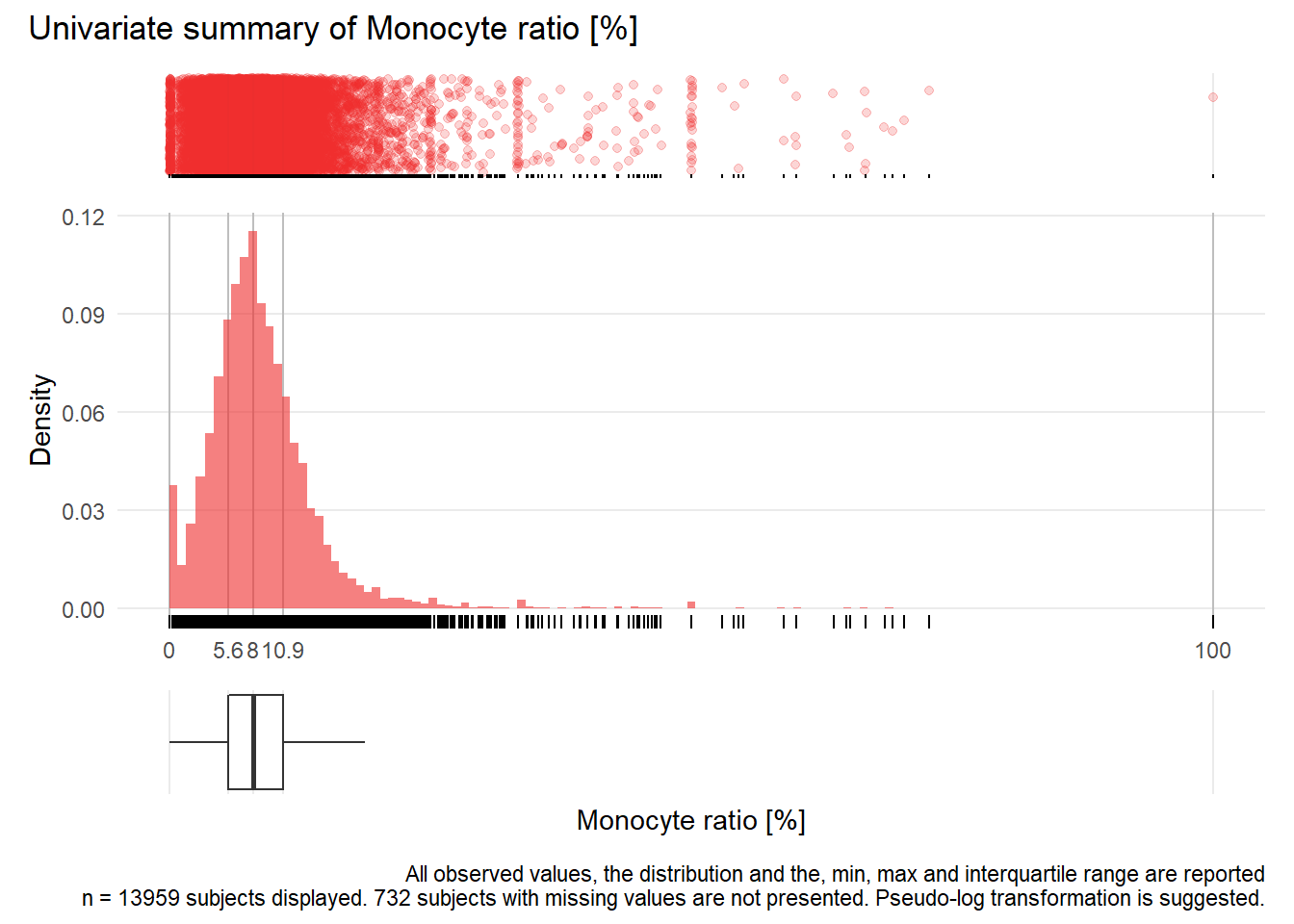
## Warning: Removed 1 rows containing missing values (geom_point).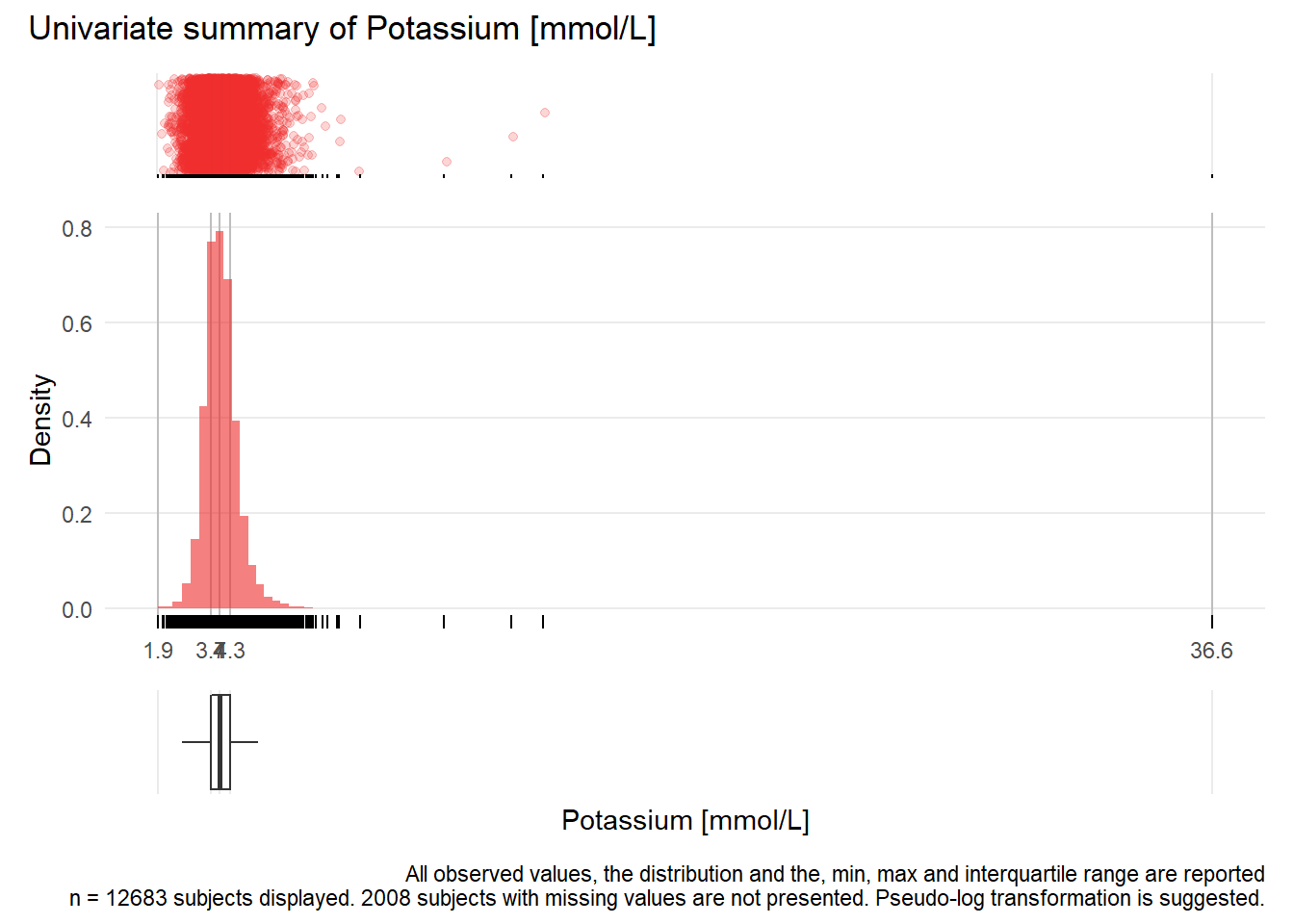
## Warning: Removed 1 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).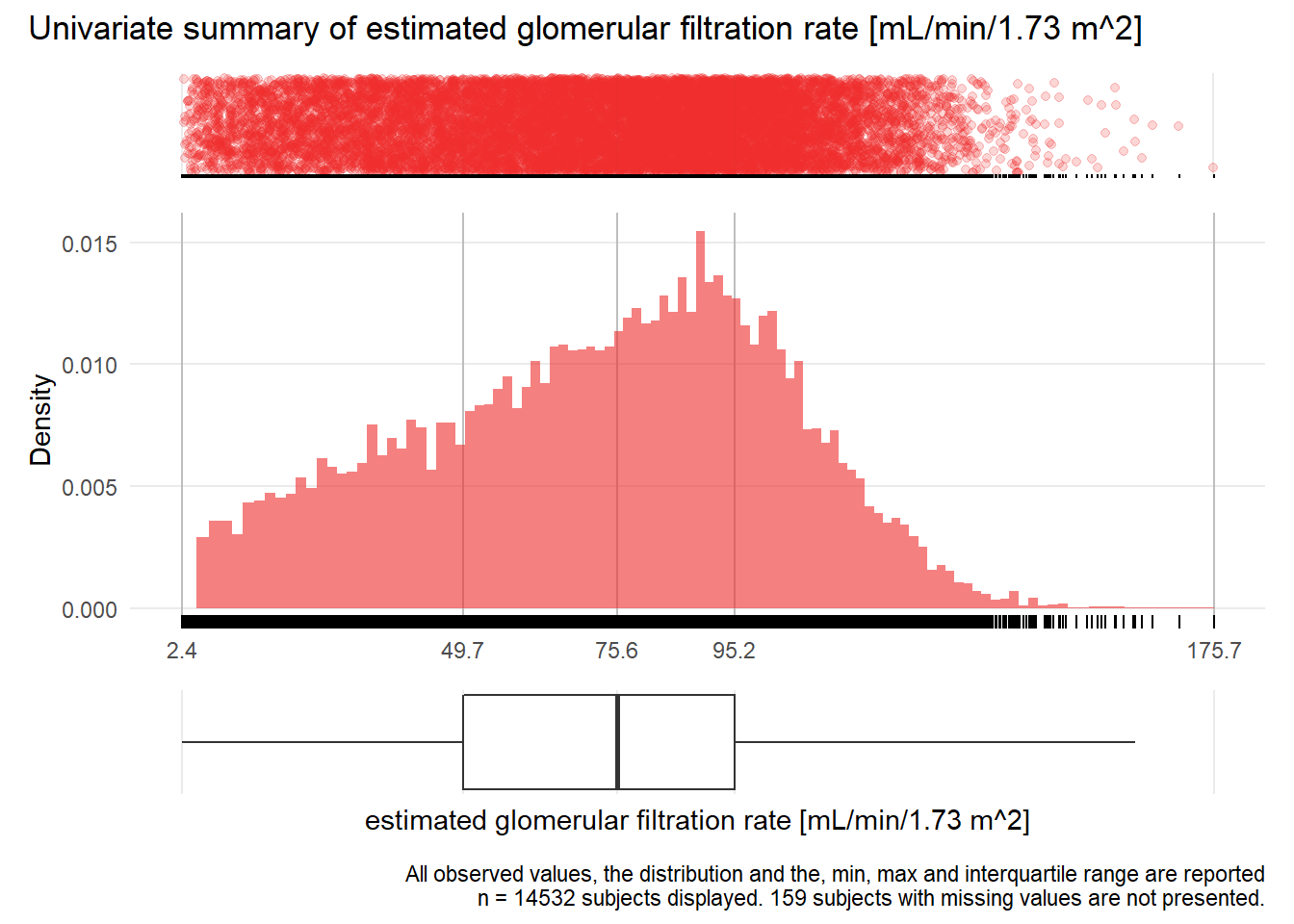
## Warning: Removed 1 rows containing missing values (geom_point).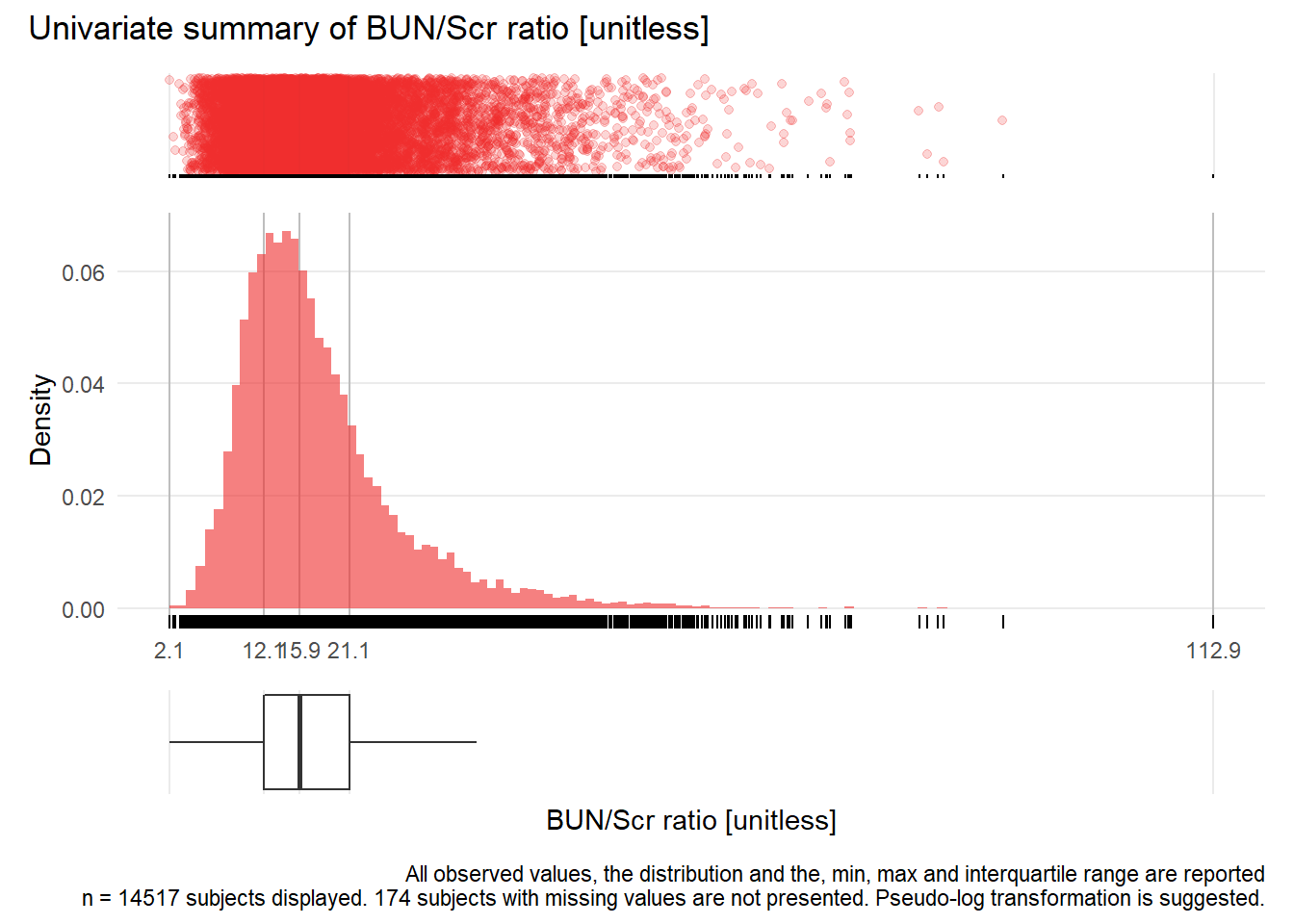
## Warning: Removed 2 rows containing missing values (geom_point).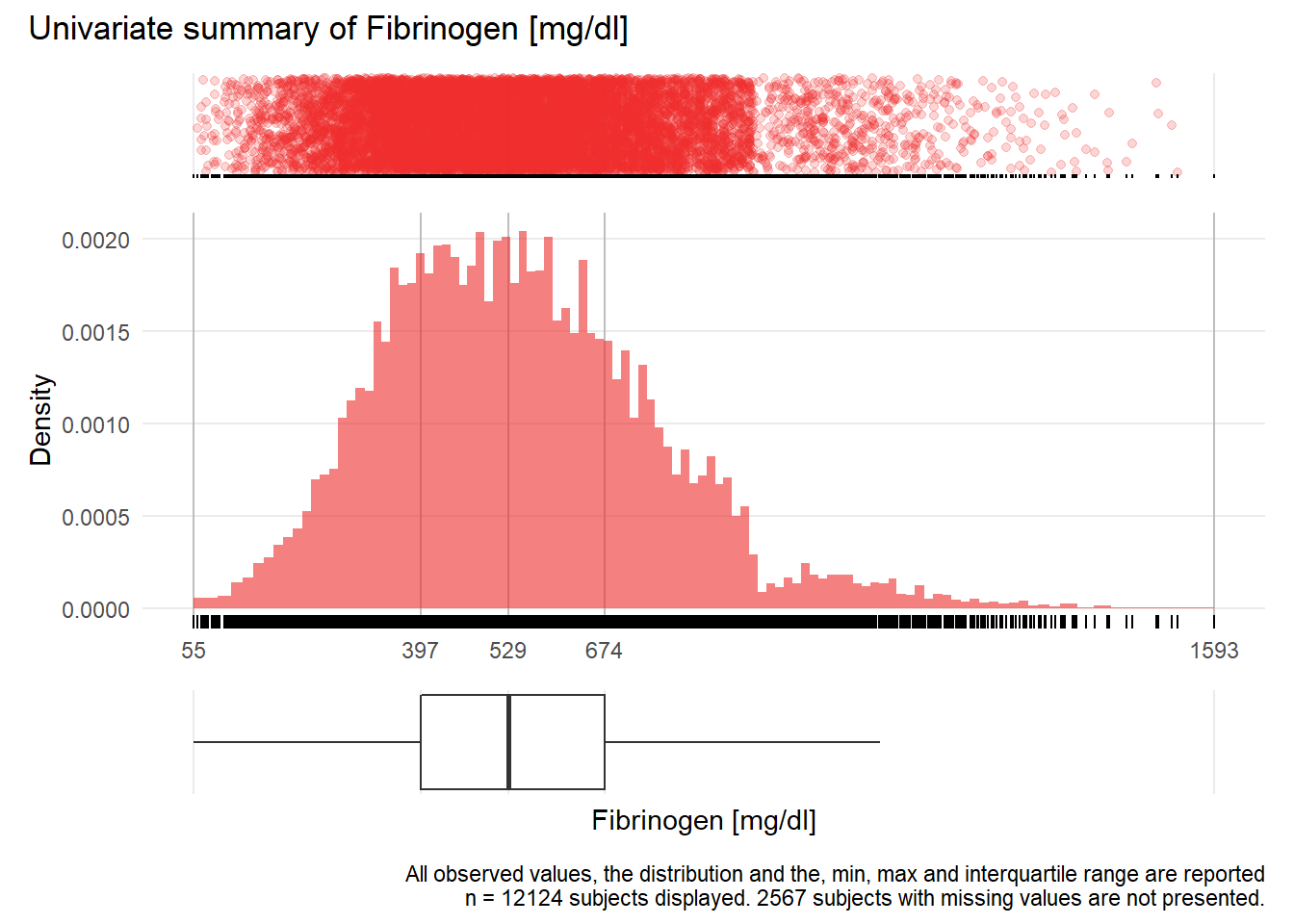
## Warning: Removed 46 rows containing missing values (geom_point).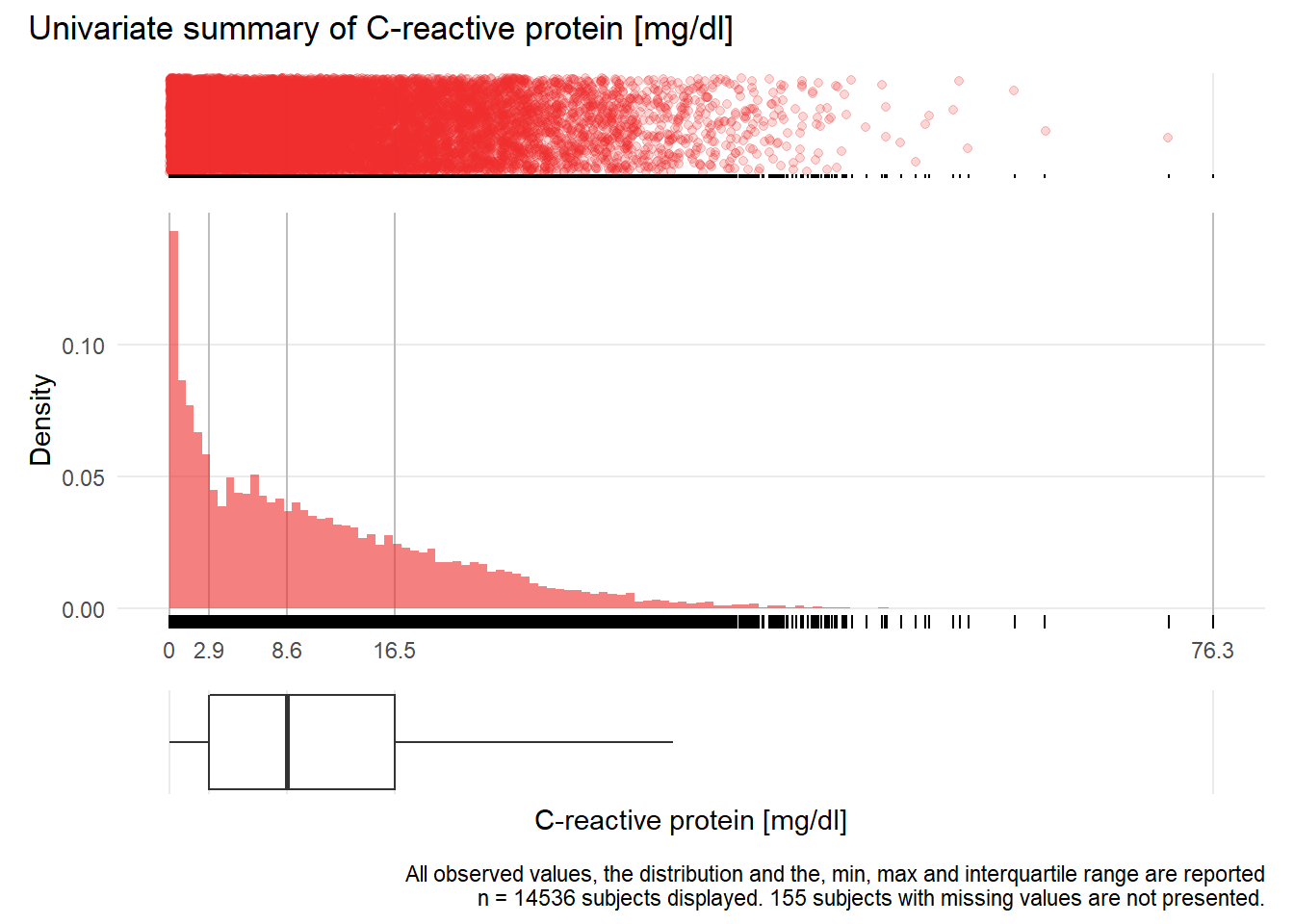
## Warning: Removed 1 rows containing missing values (geom_point).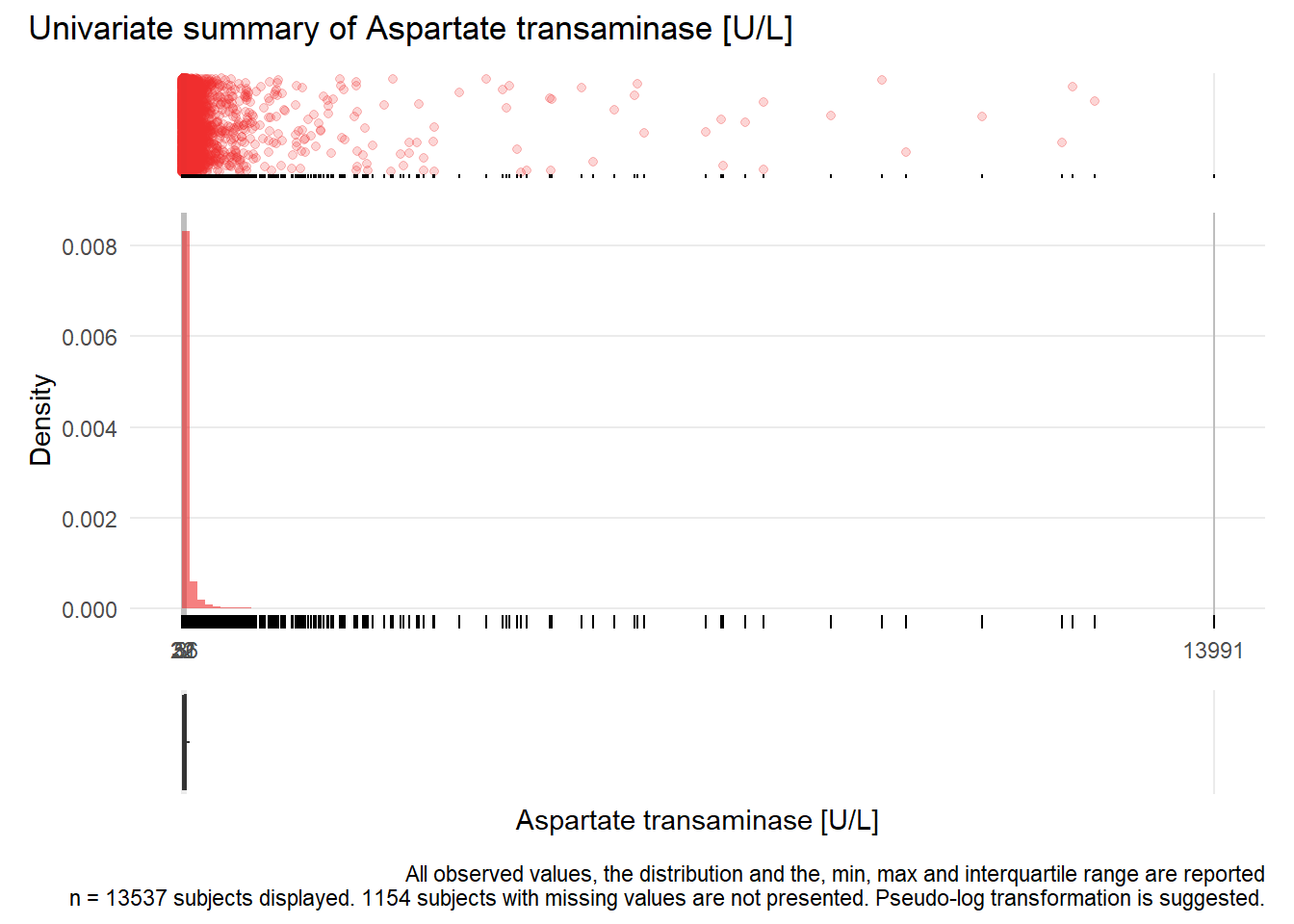
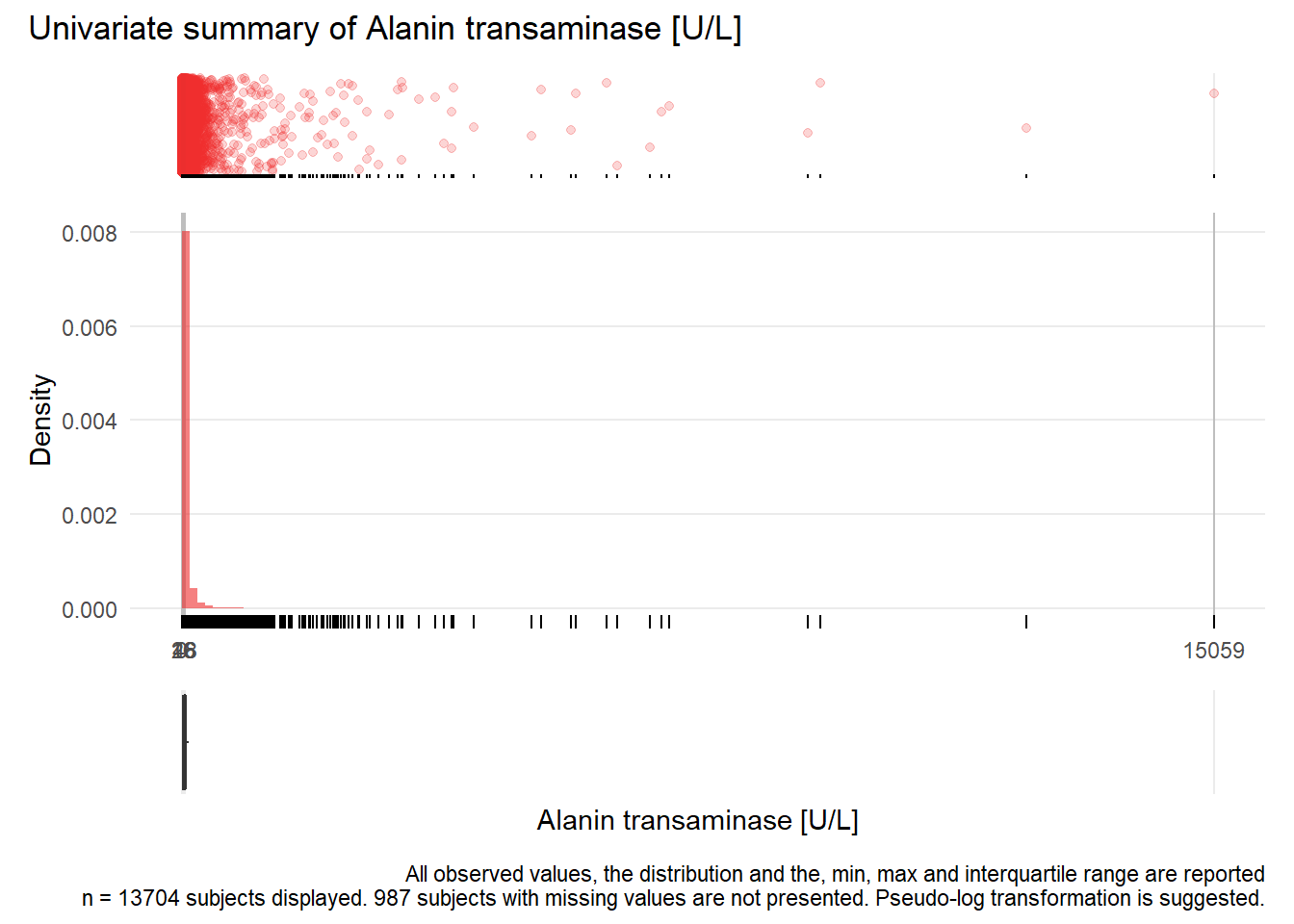
## Warning: Removed 1 rows containing missing values (geom_point).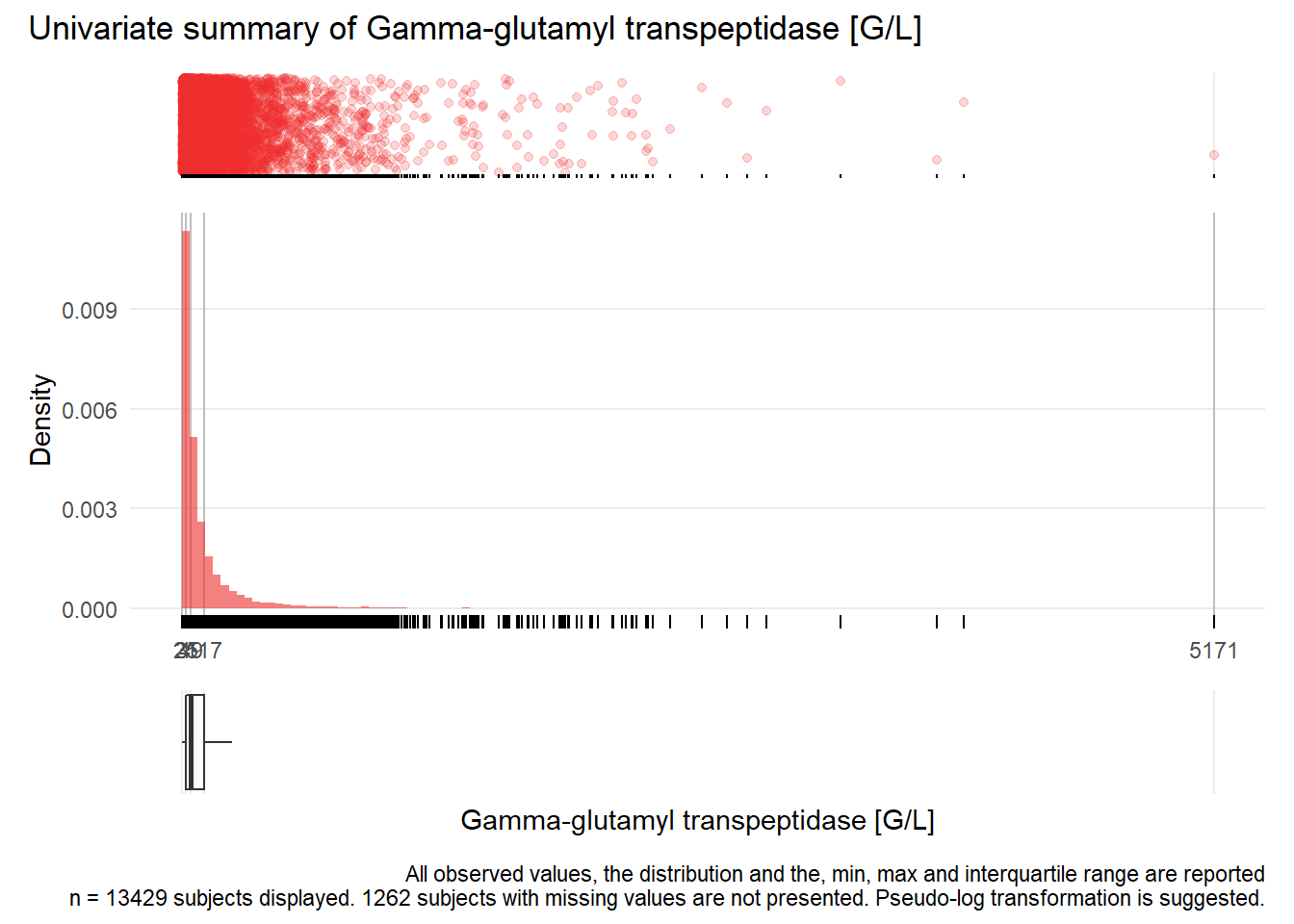
## Warning: Removed 2 rows containing missing values (geom_point).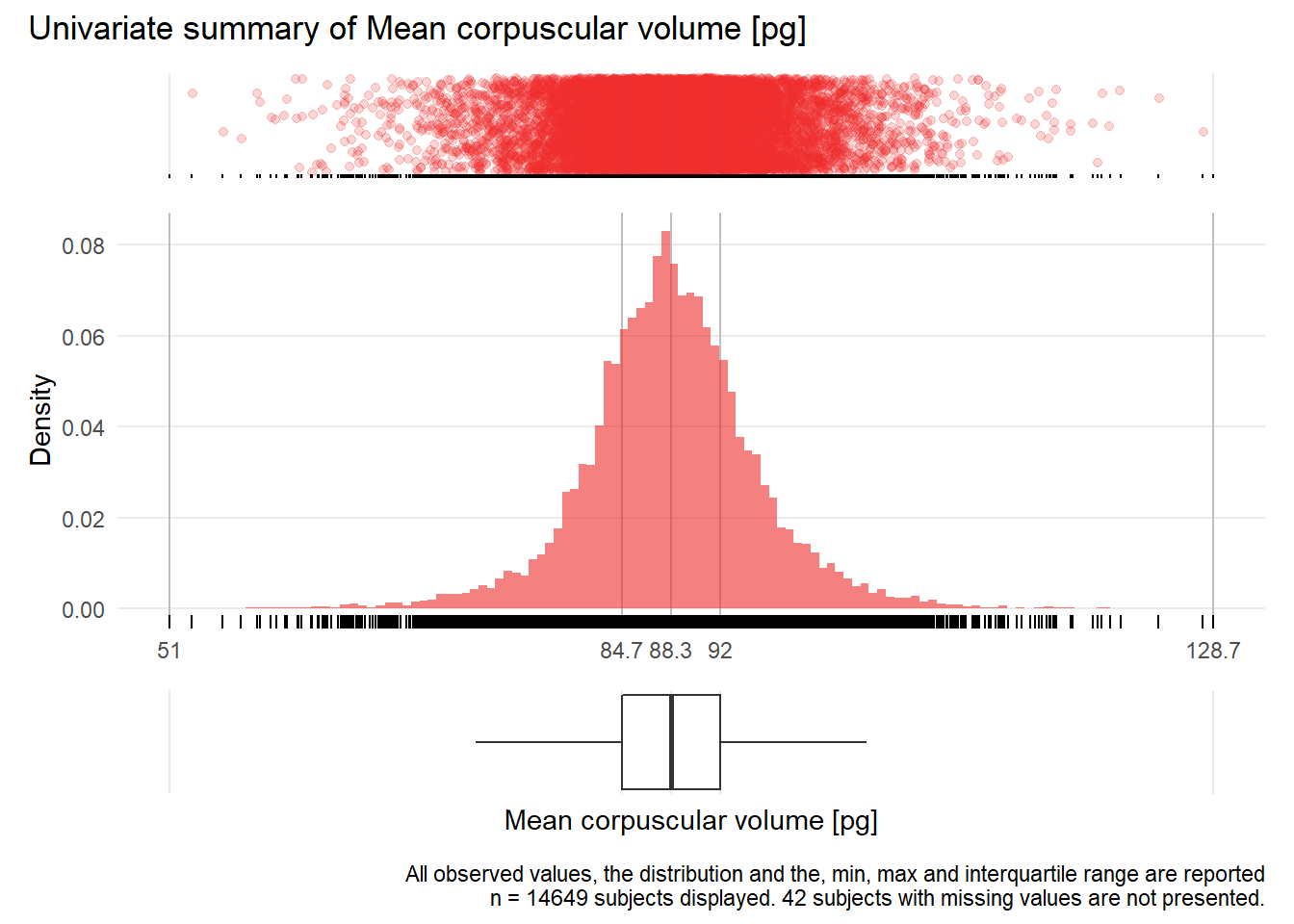
## Warning: Removed 7 rows containing missing values (geom_point).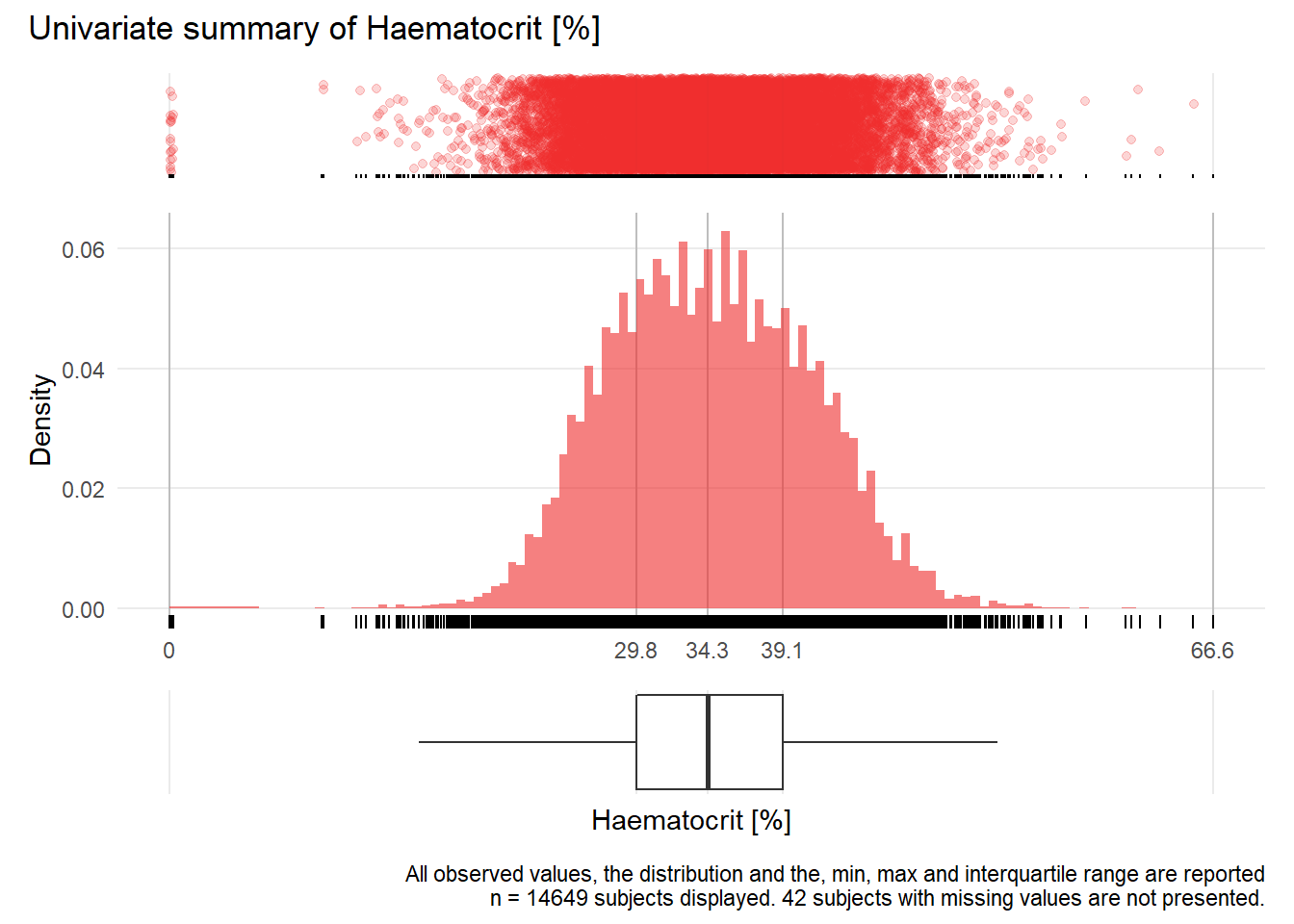
## Warning: Removed 2 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).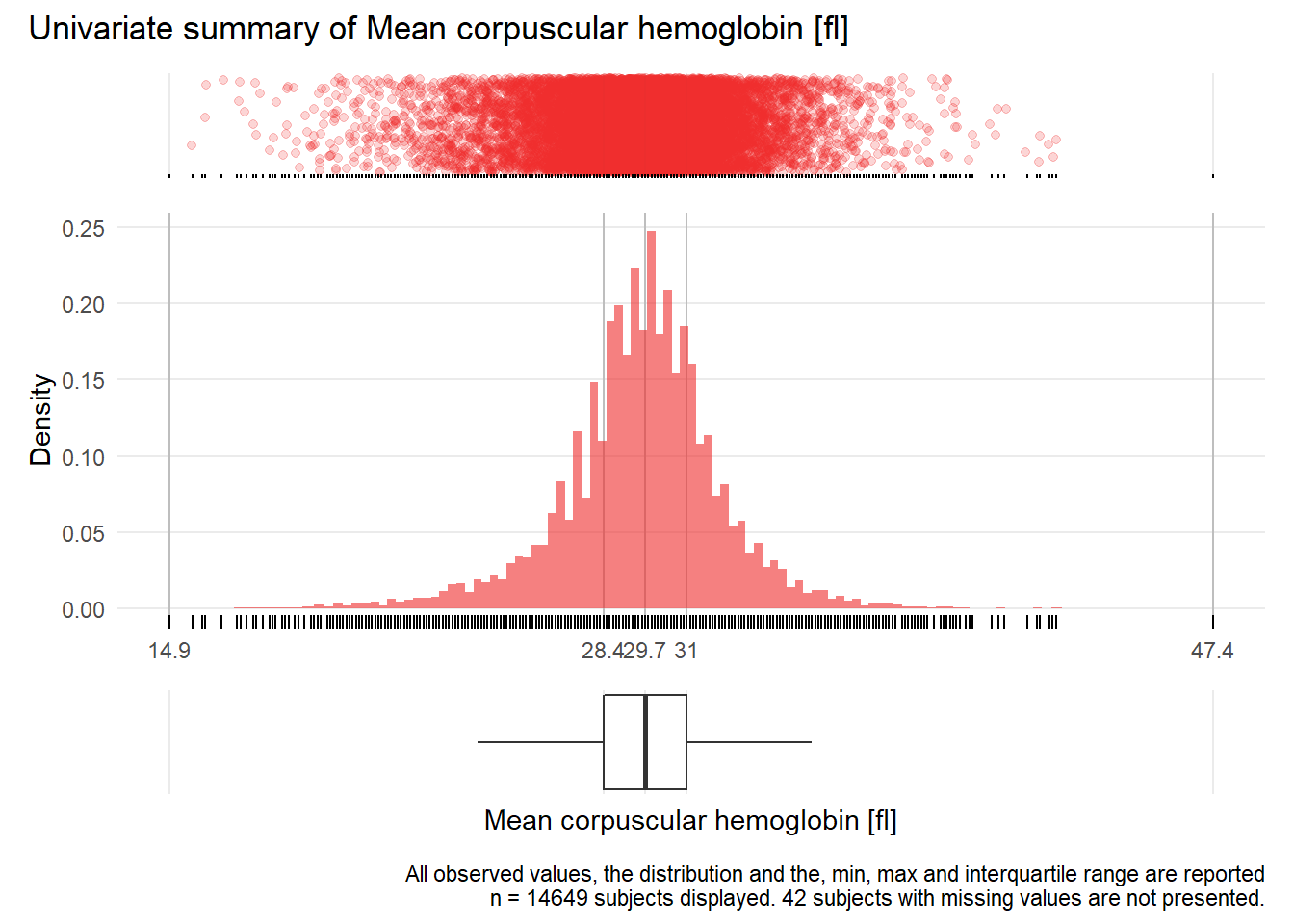
## Warning: Removed 1 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).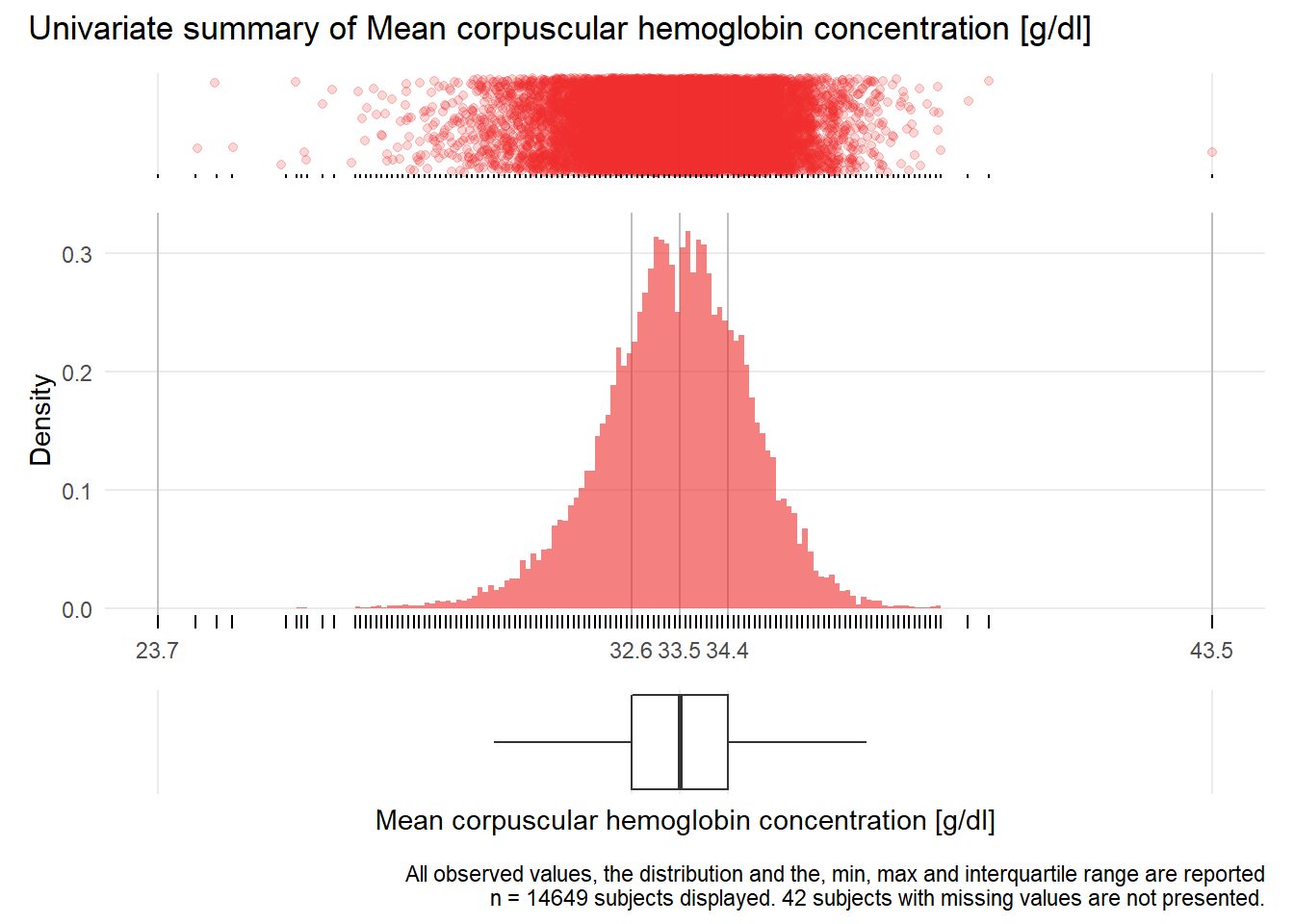
## Warning: Removed 1 rows containing missing values (geom_point).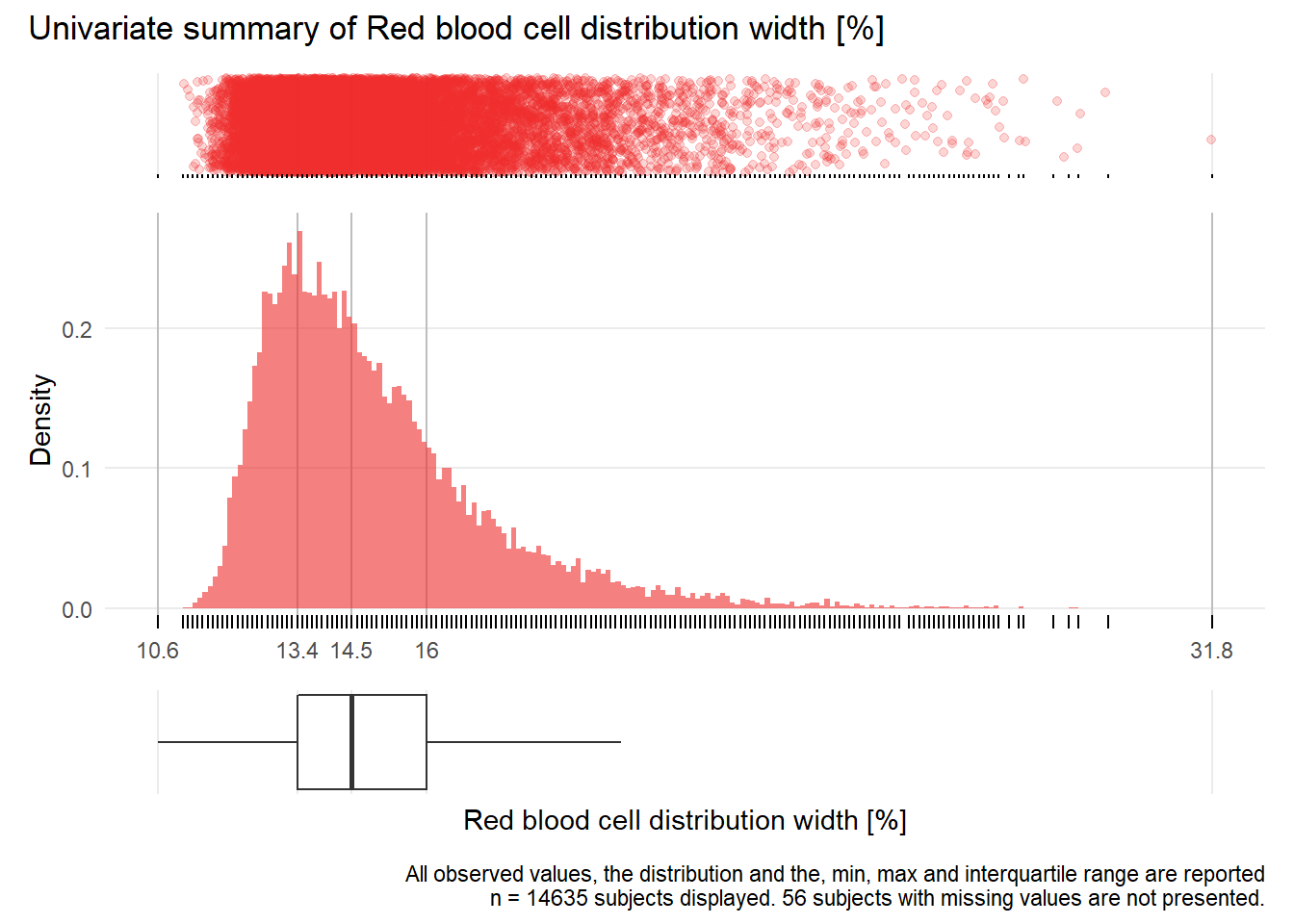
## Warning: Removed 2 rows containing missing values (geom_point).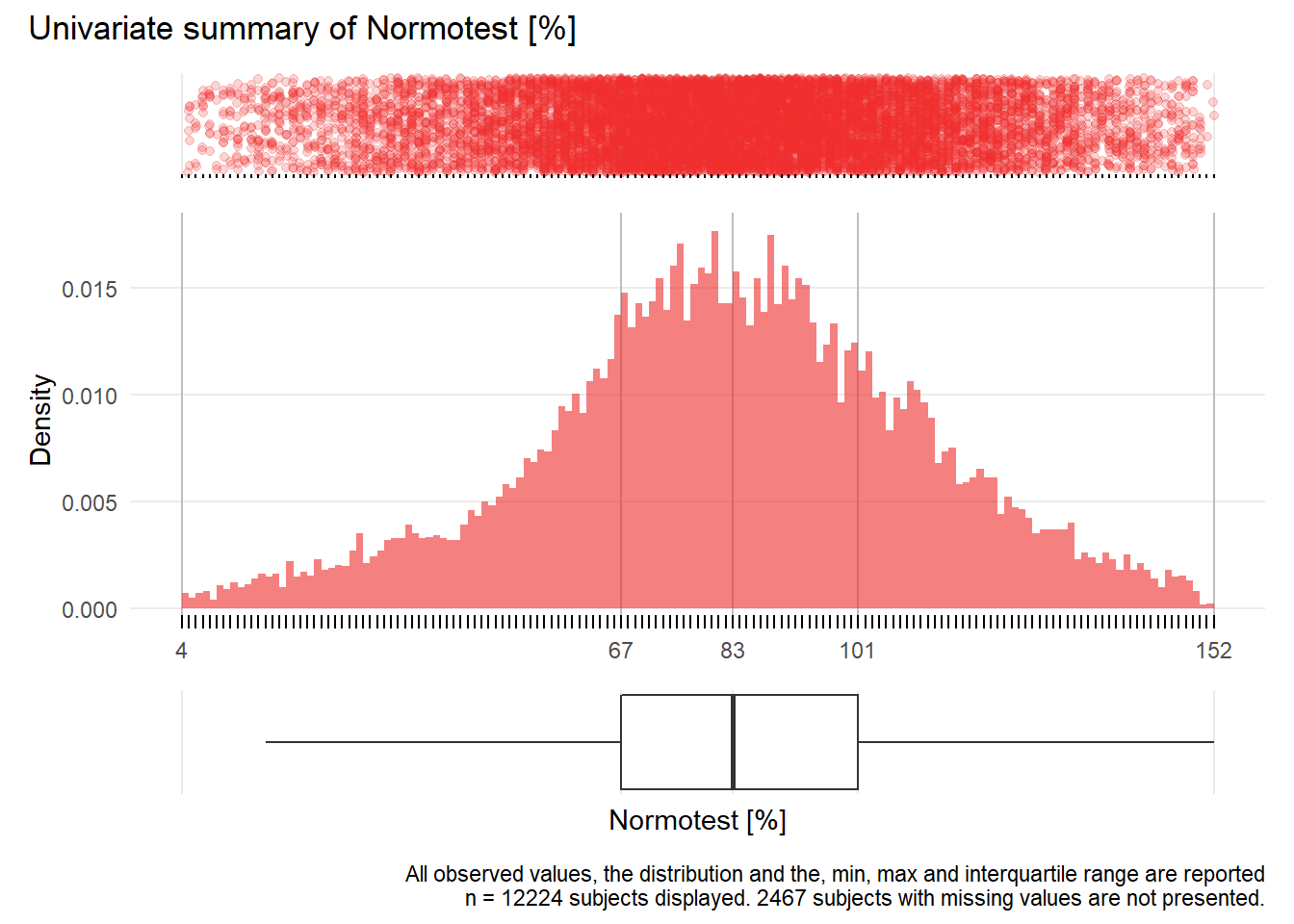
## Warning: Removed 1 rows containing missing values (geom_point).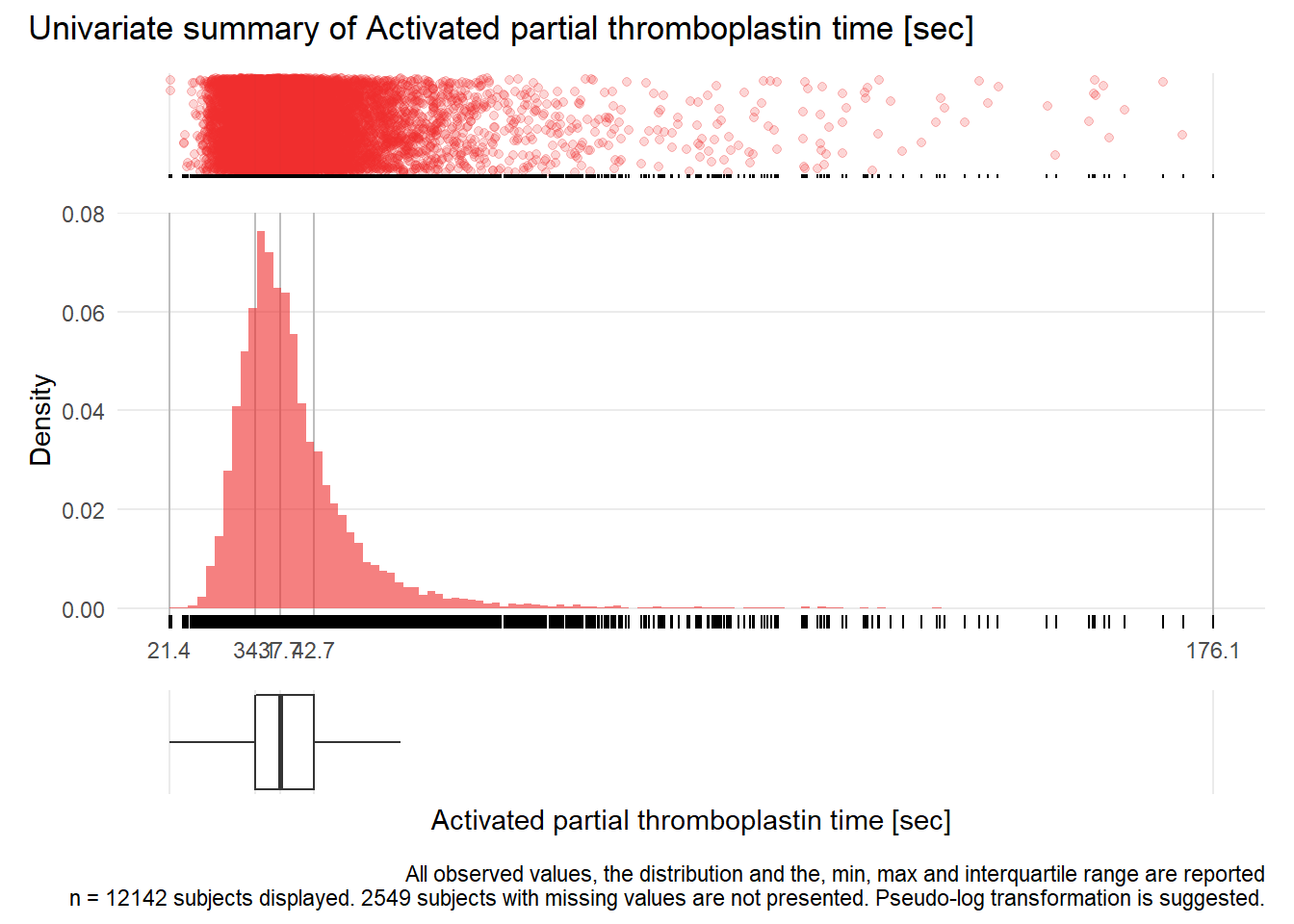
## Warning: Removed 1 rows containing missing values (geom_bar).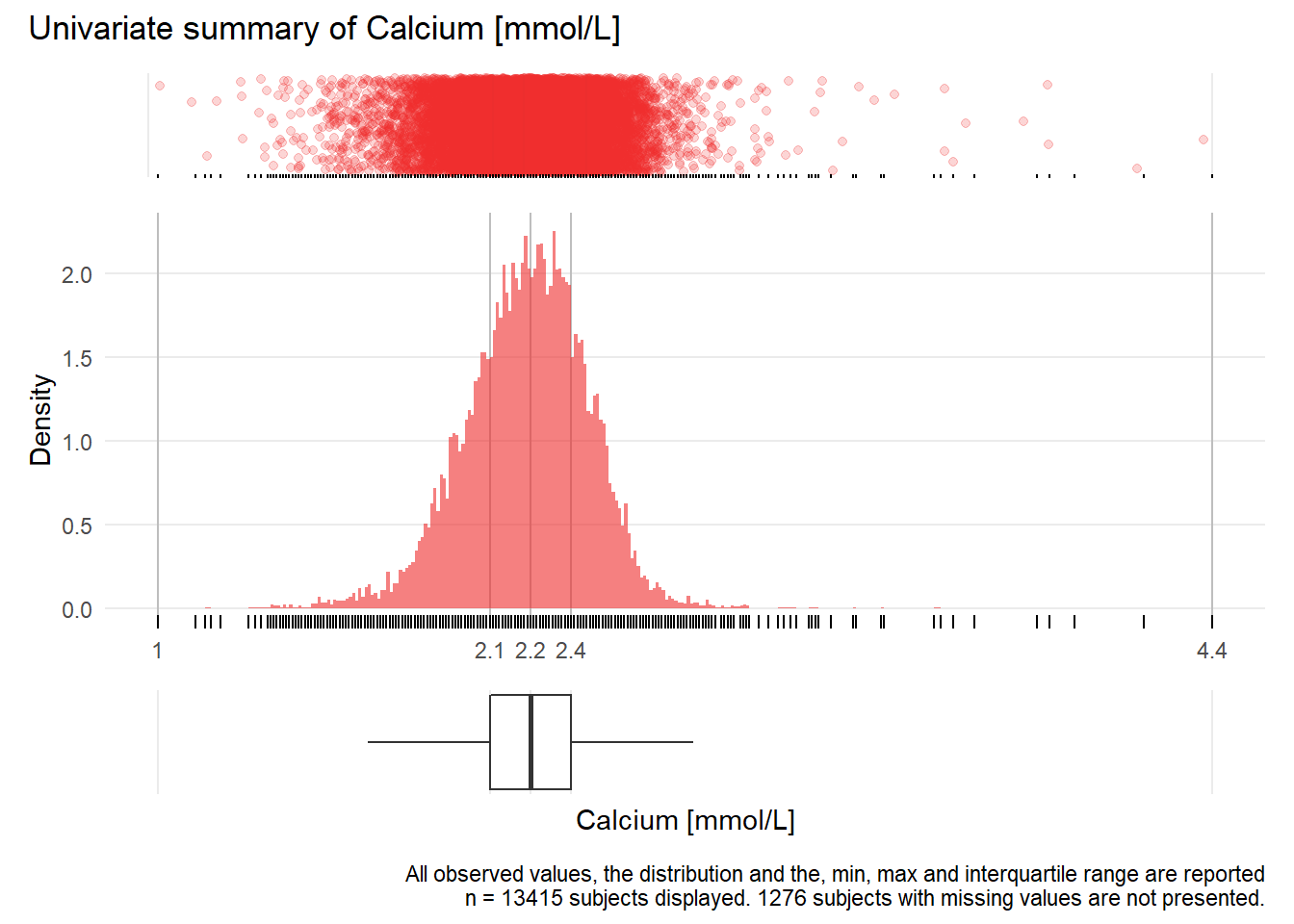
## Warning: Removed 28 rows containing missing values (geom_point).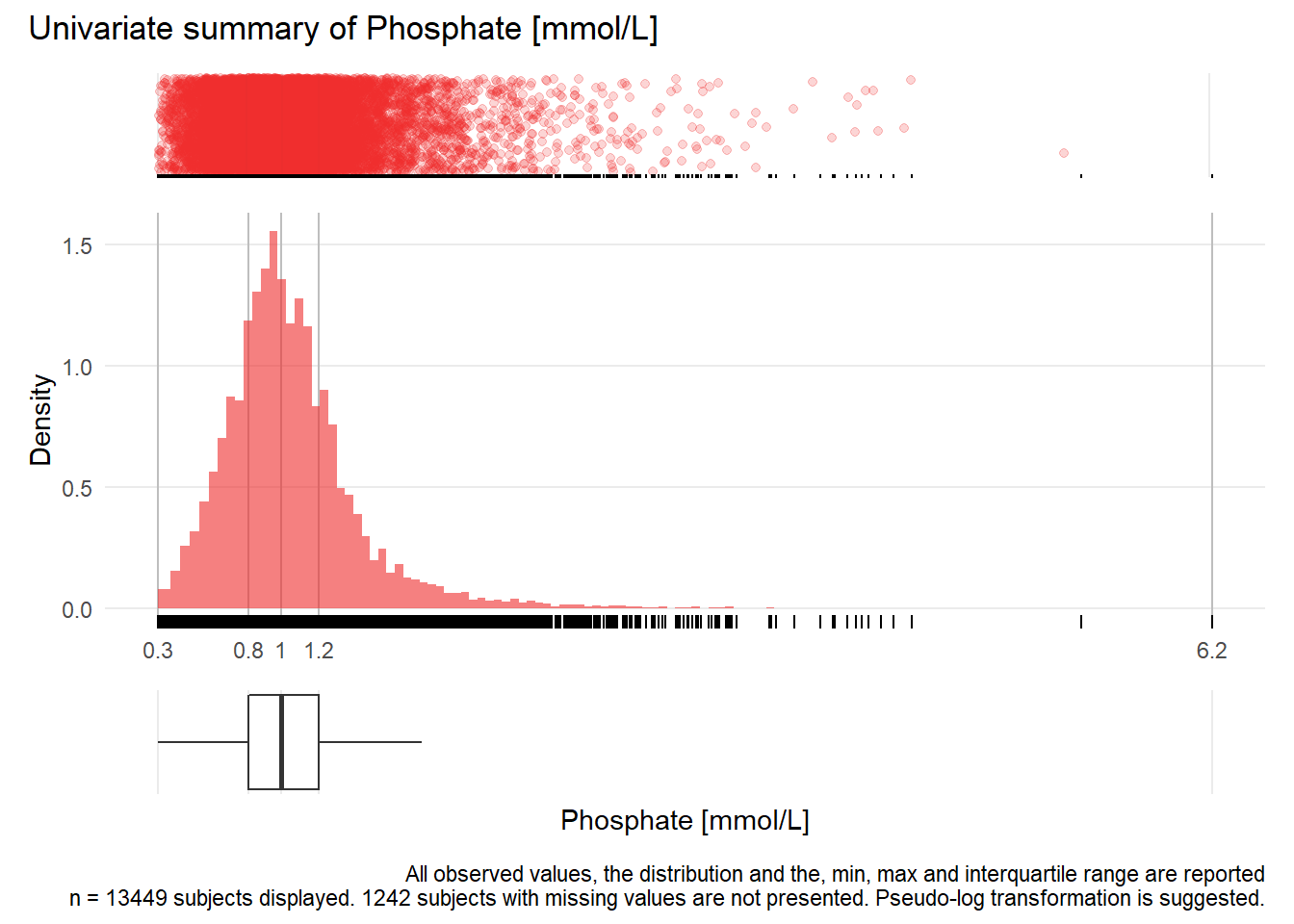
## Warning: Removed 1 rows containing missing values (geom_point).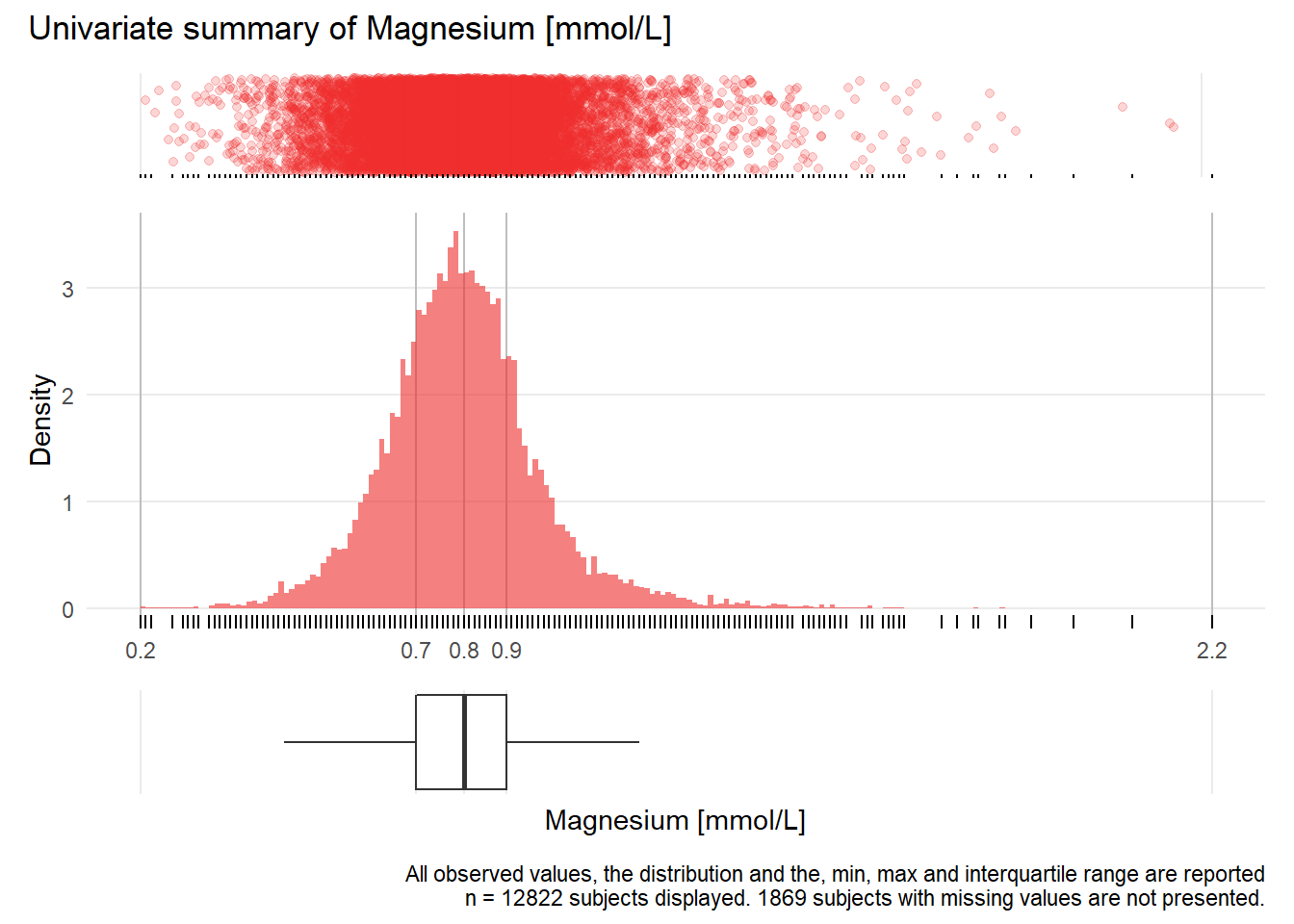
## Warning: Removed 1 rows containing missing values (geom_point).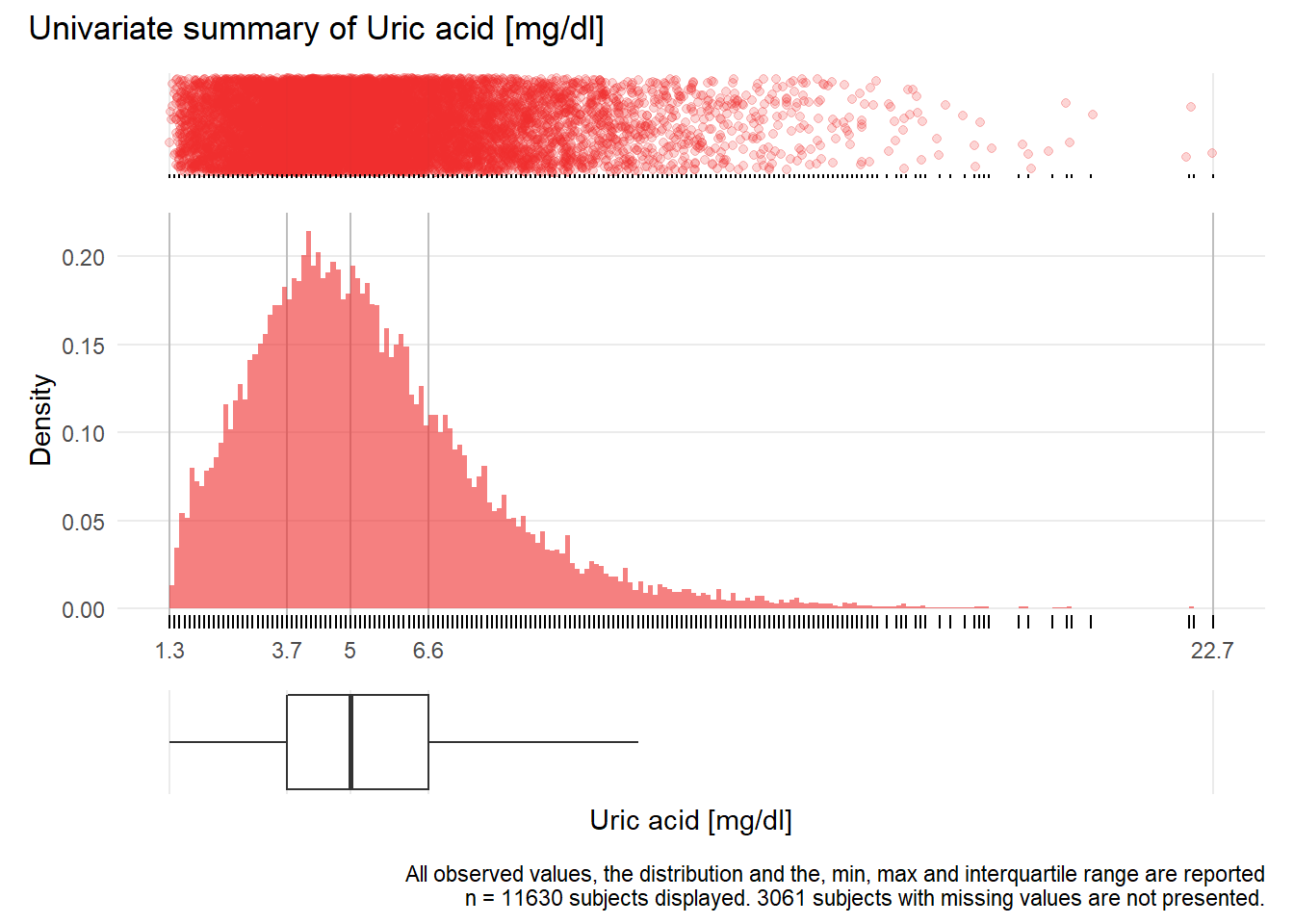
## Warning: Removed 5 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).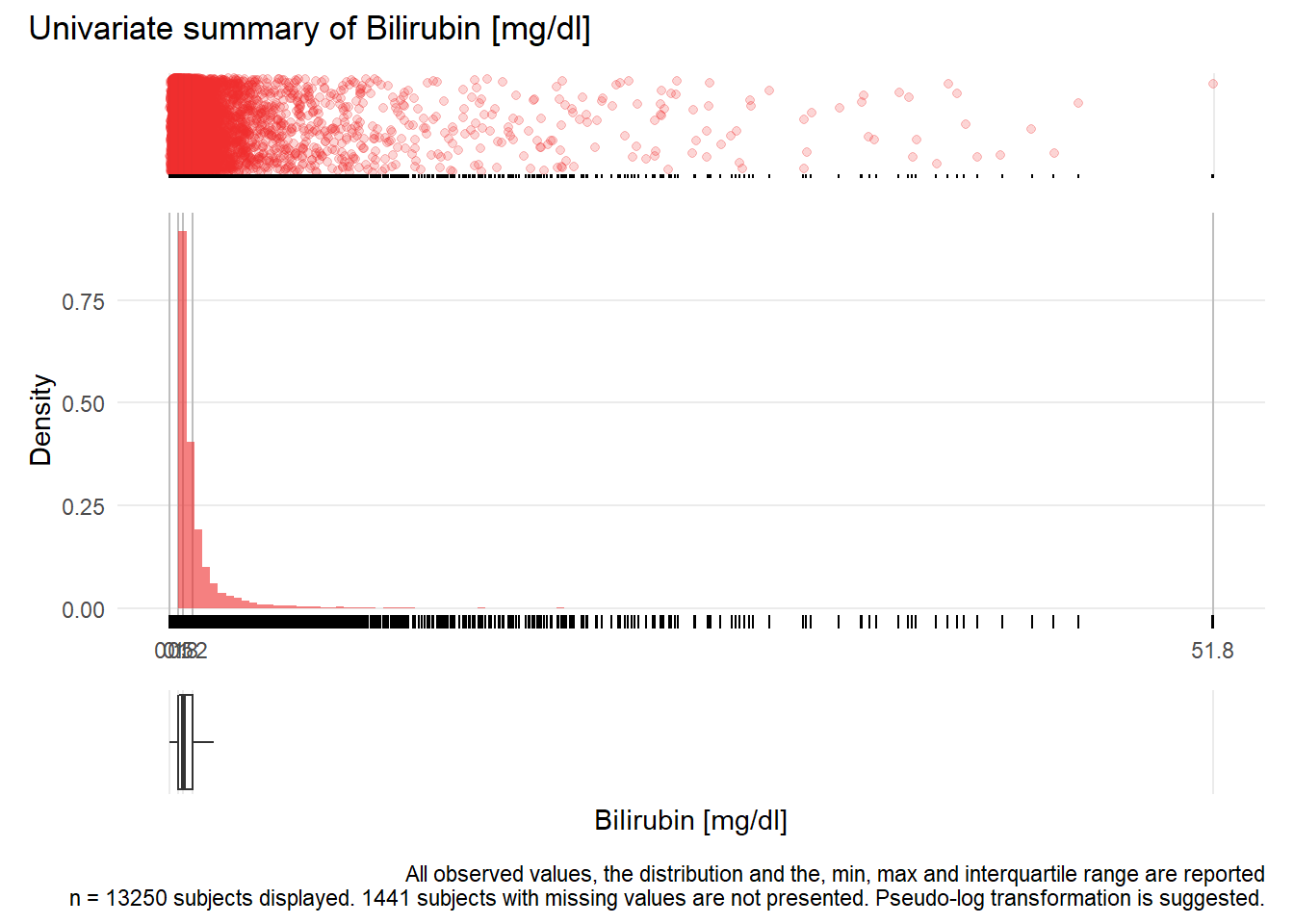
## Warning: Removed 1 rows containing missing values (geom_point).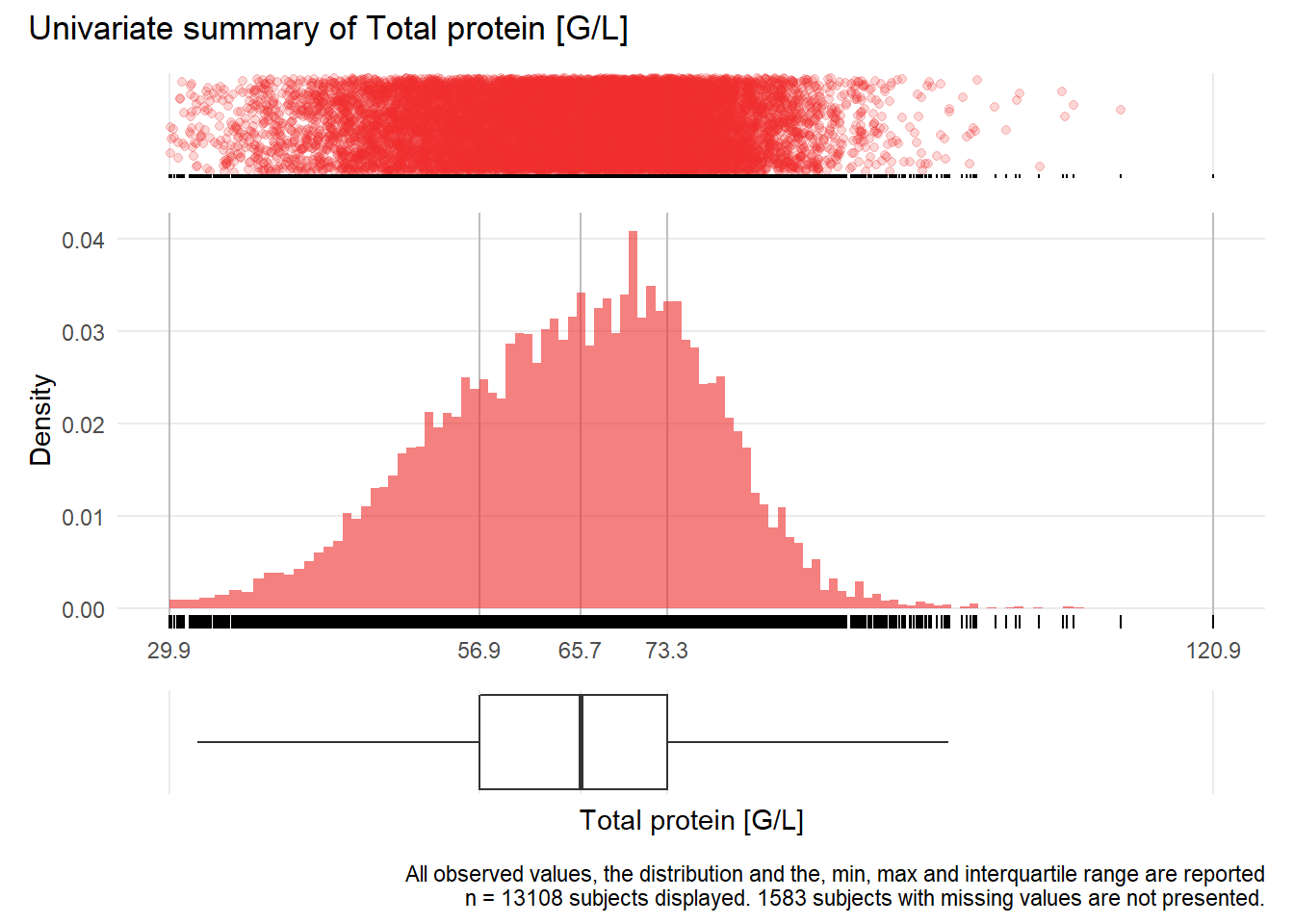
## Warning: Removed 1 rows containing missing values (geom_point).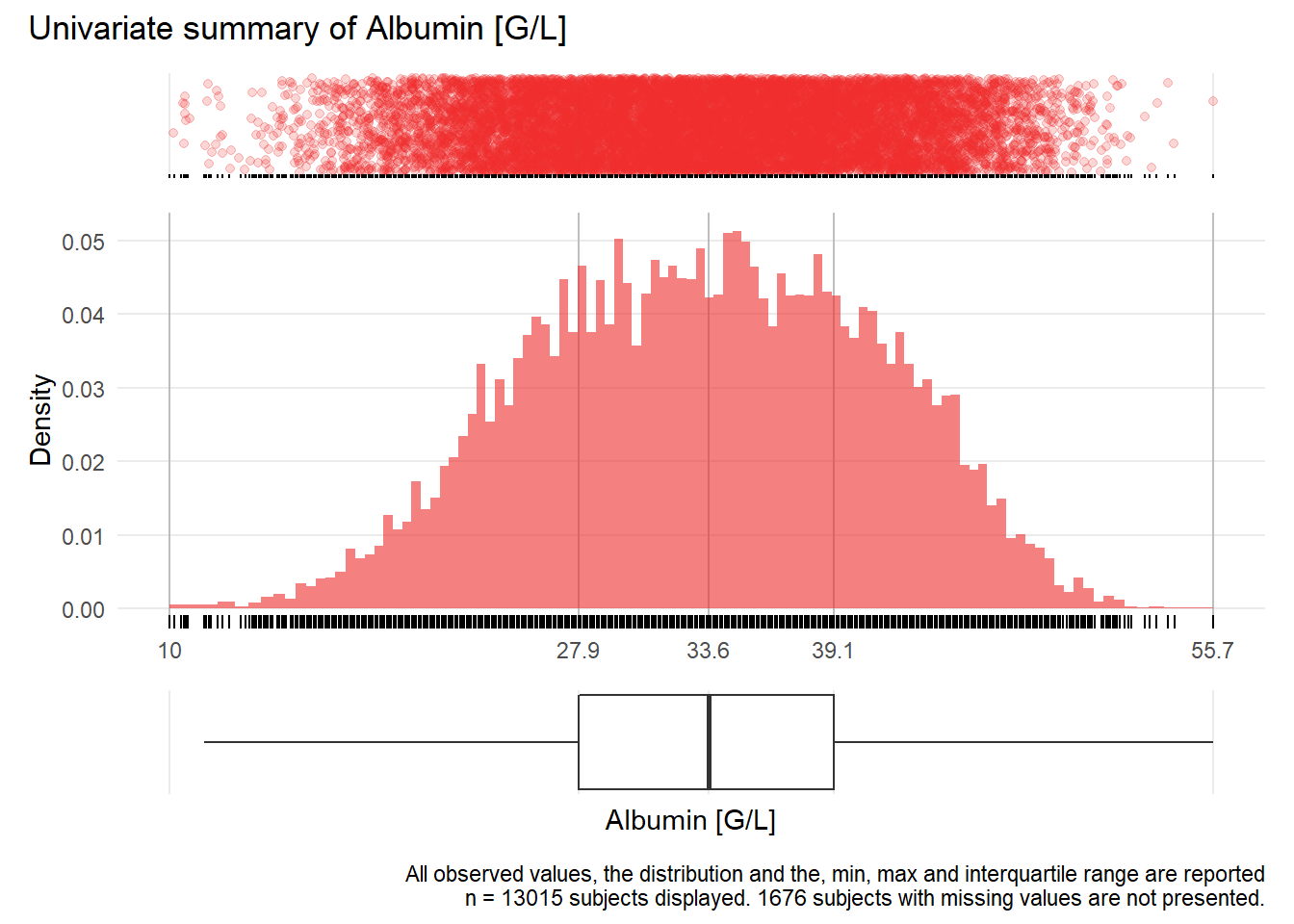
## Warning: Removed 3 rows containing missing values (geom_point).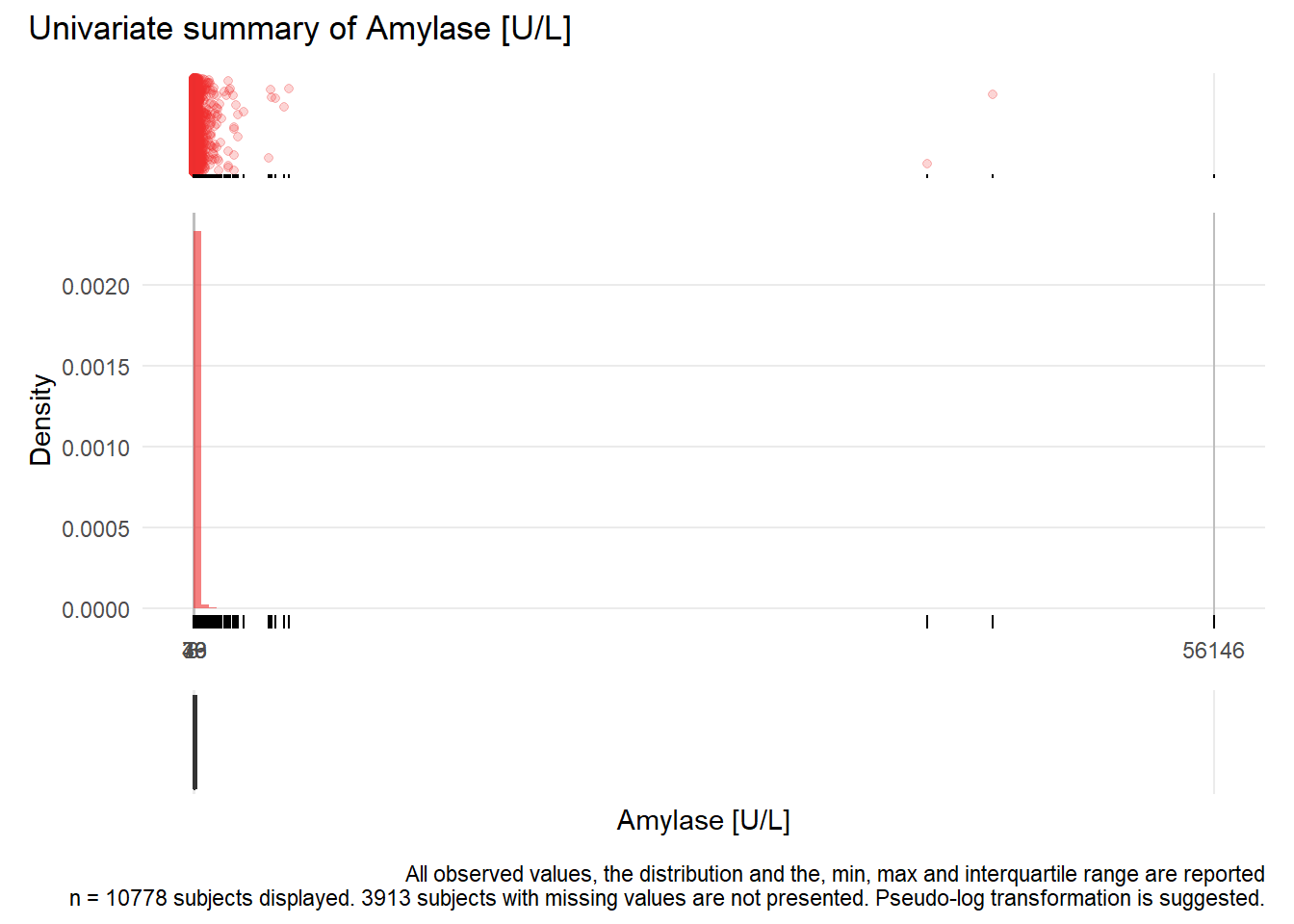
## Warning: Removed 3 rows containing missing values (geom_point).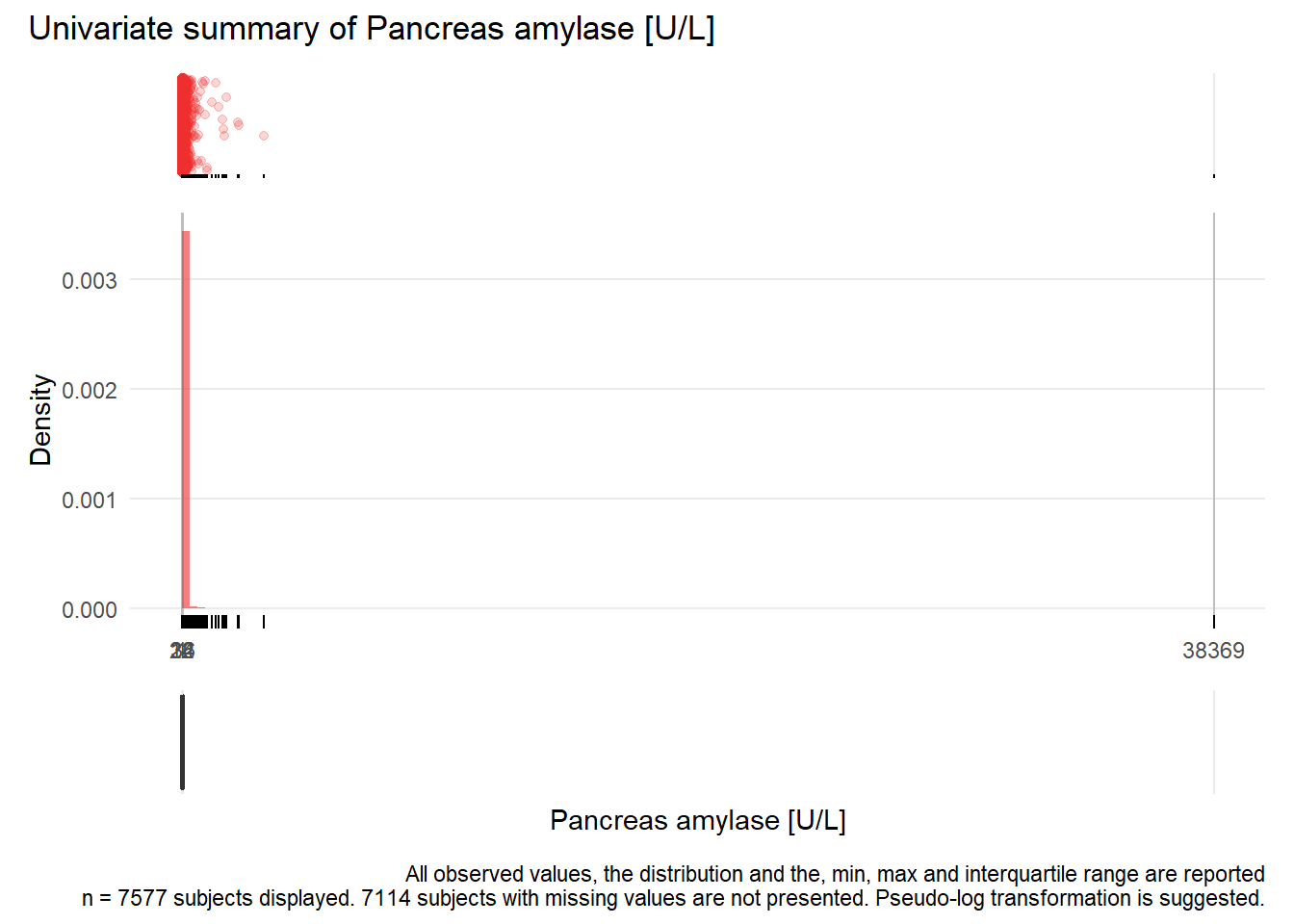
## Warning: Removed 1 rows containing missing values (geom_point).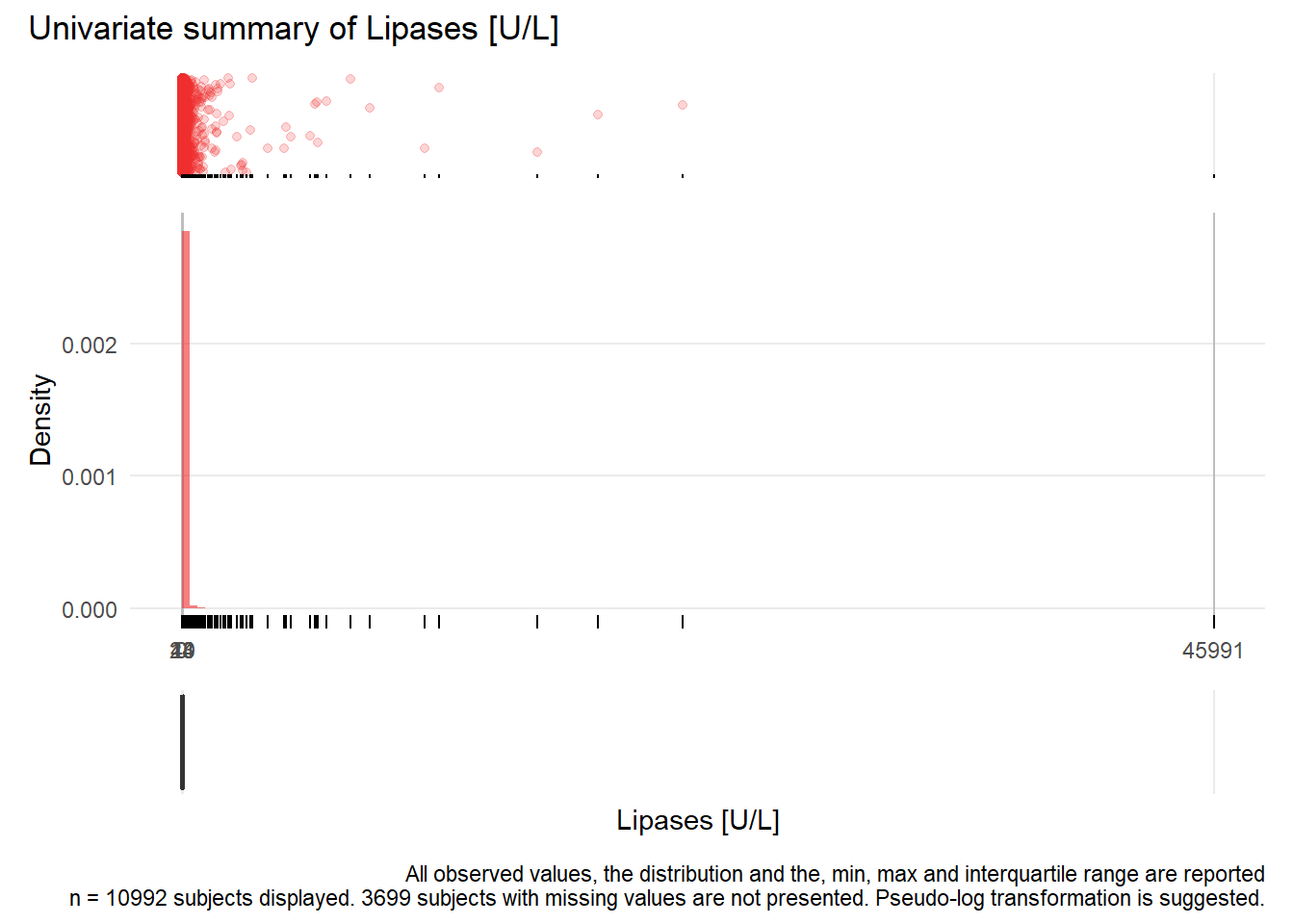
## Warning: Removed 18 rows containing missing values (geom_point).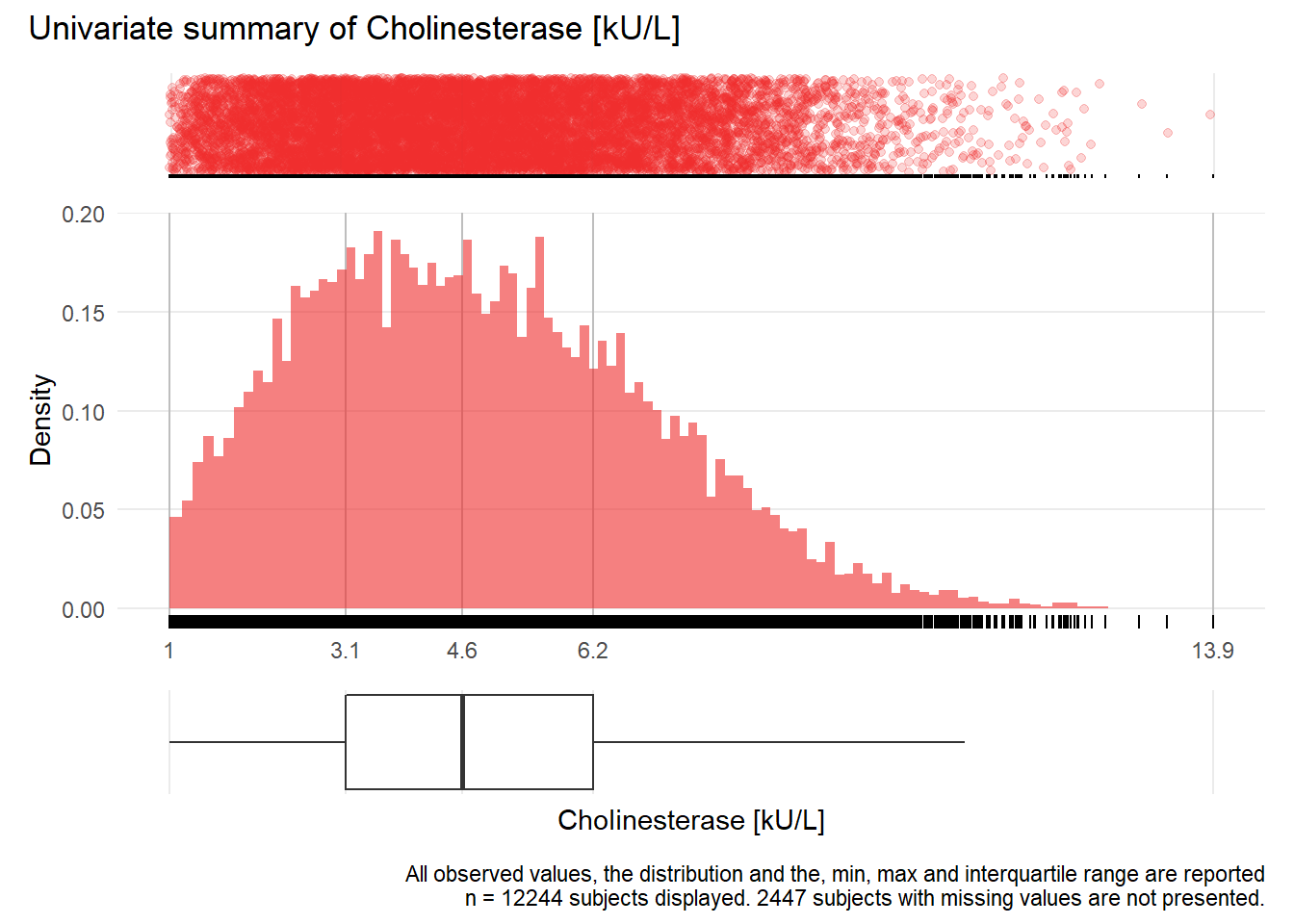
## Warning: Removed 2 rows containing missing values (geom_point).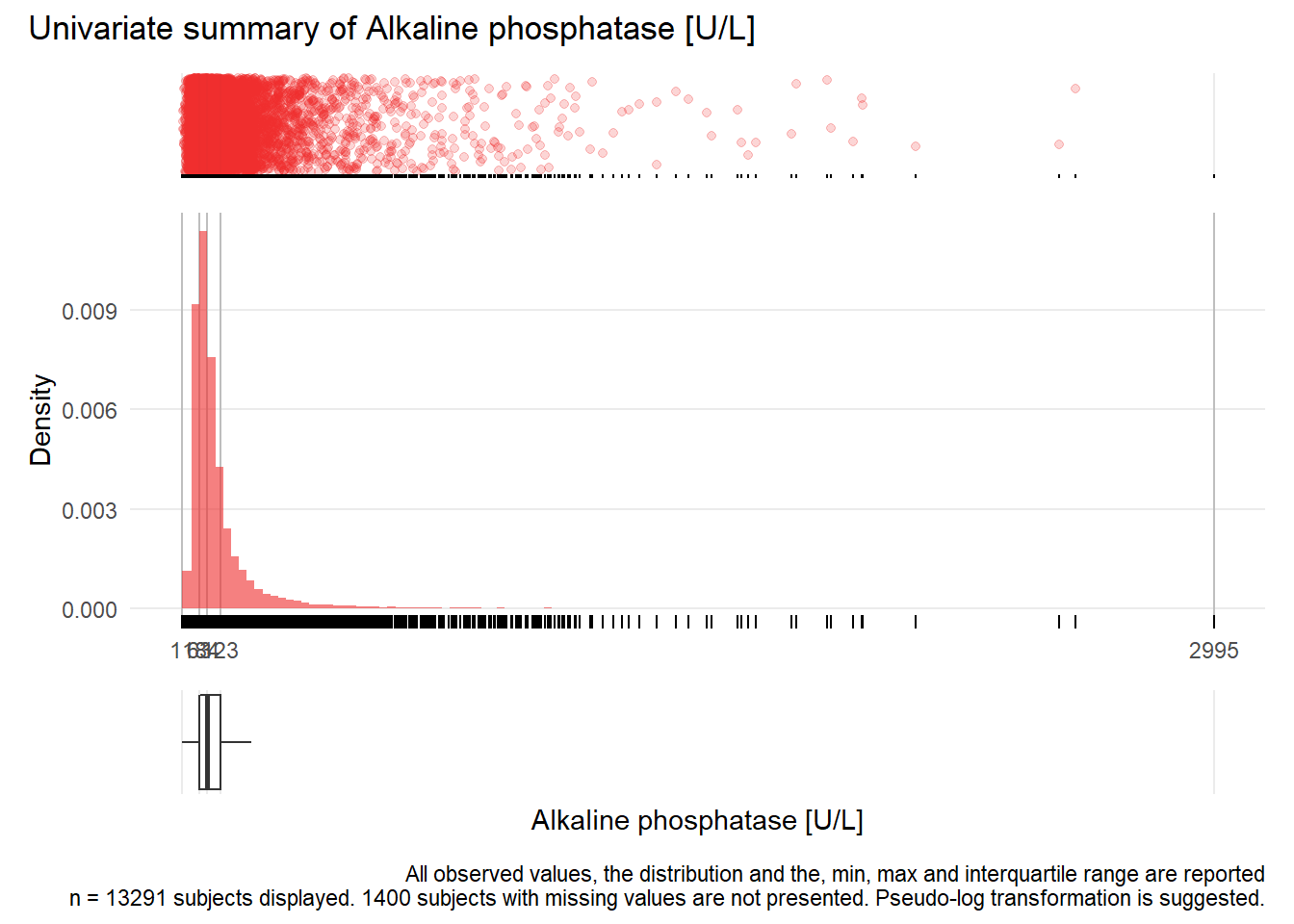
## Warning: Removed 1 rows containing missing values (geom_point).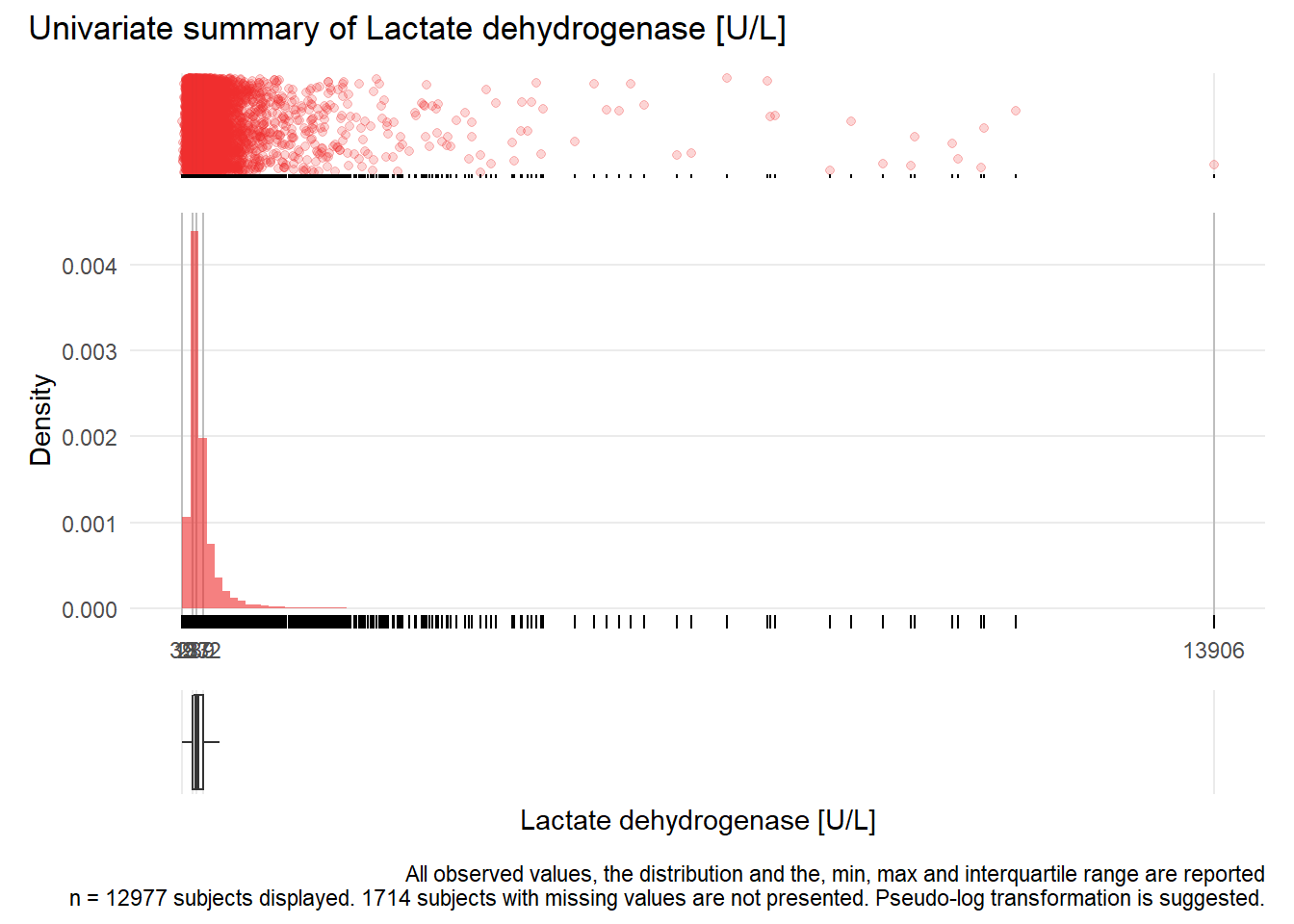
## Warning: Removed 2 rows containing missing values (geom_point).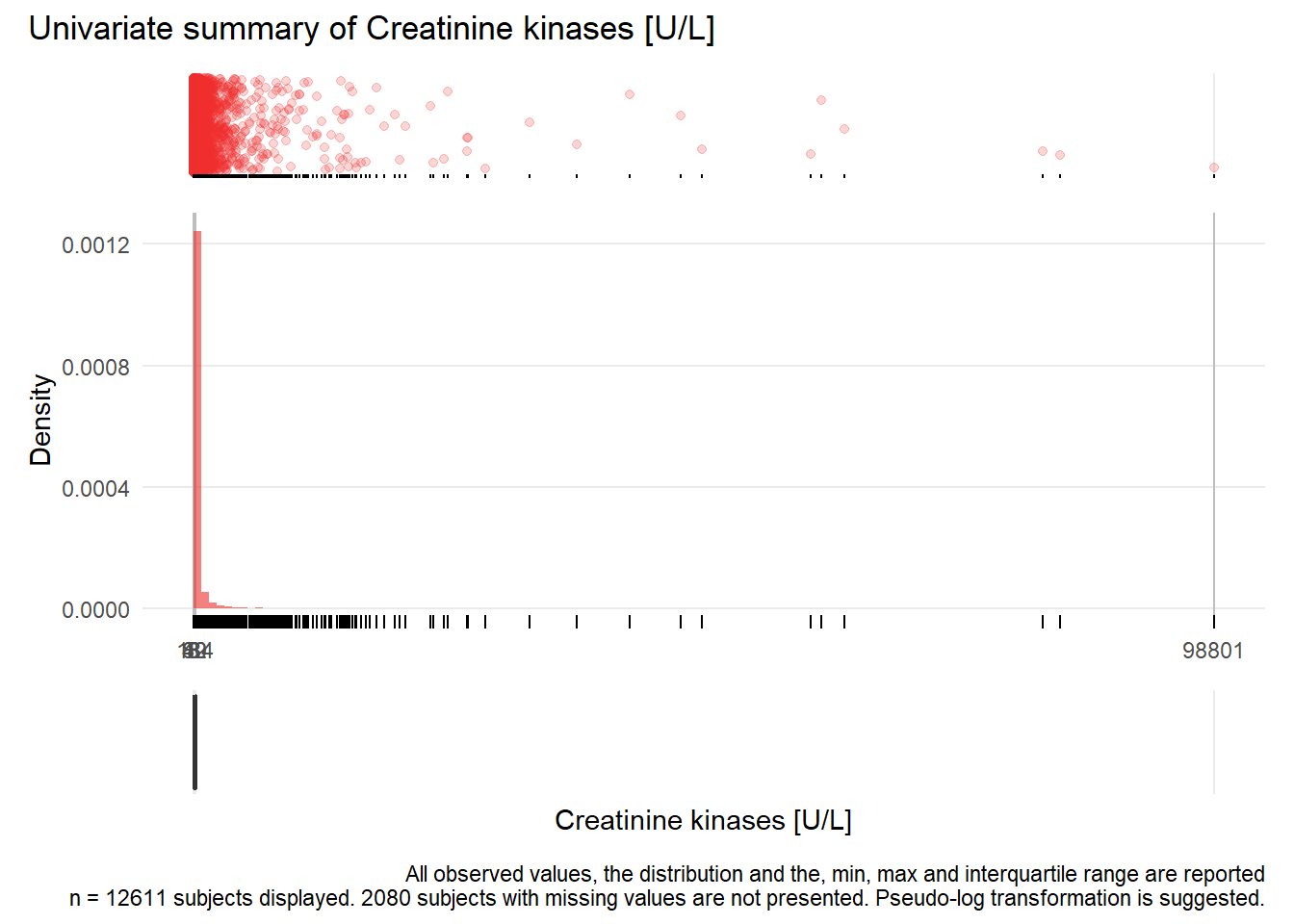
## Warning: Removed 2 rows containing missing values (geom_point).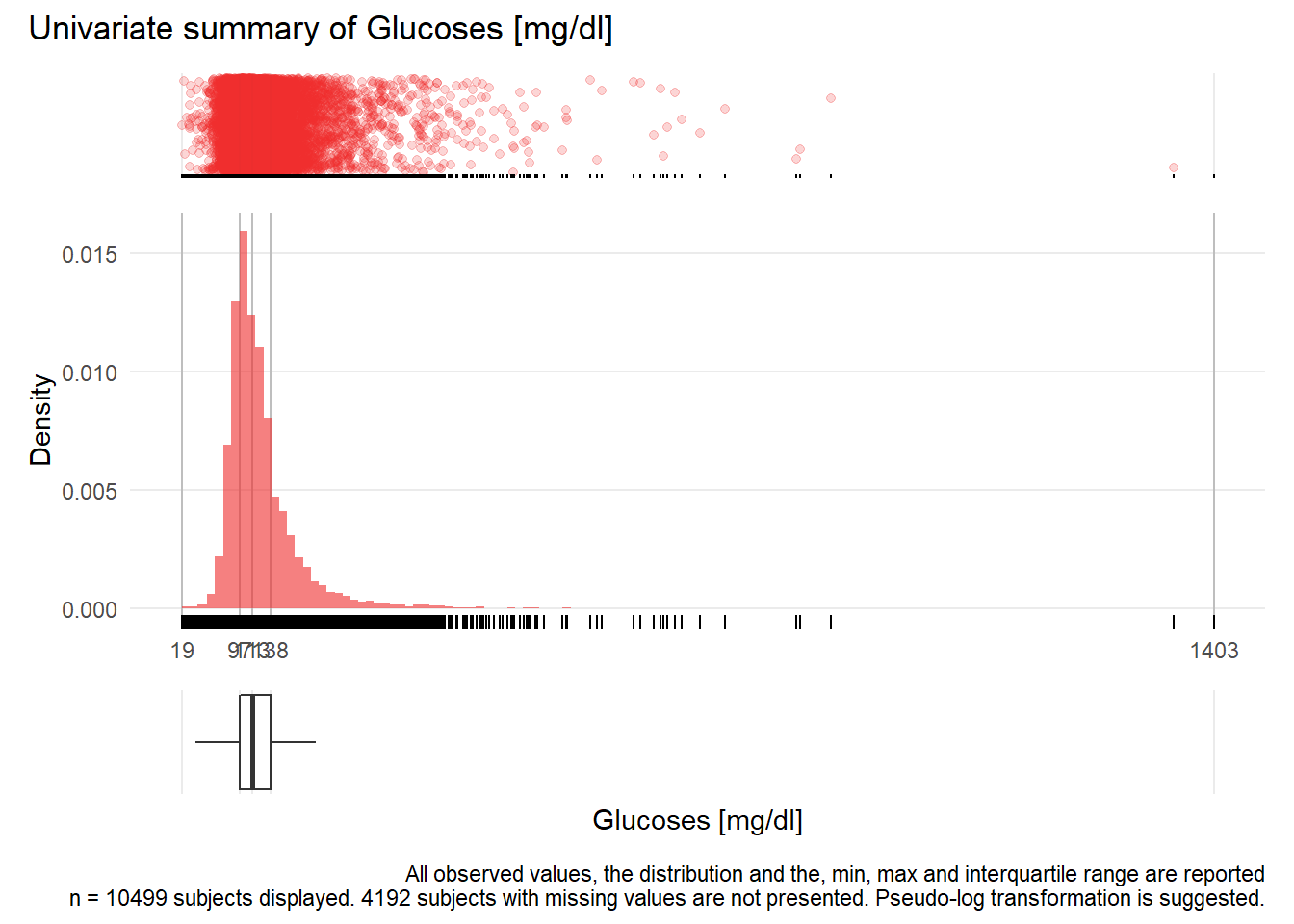
## Warning: Removed 1 rows containing missing values (geom_point).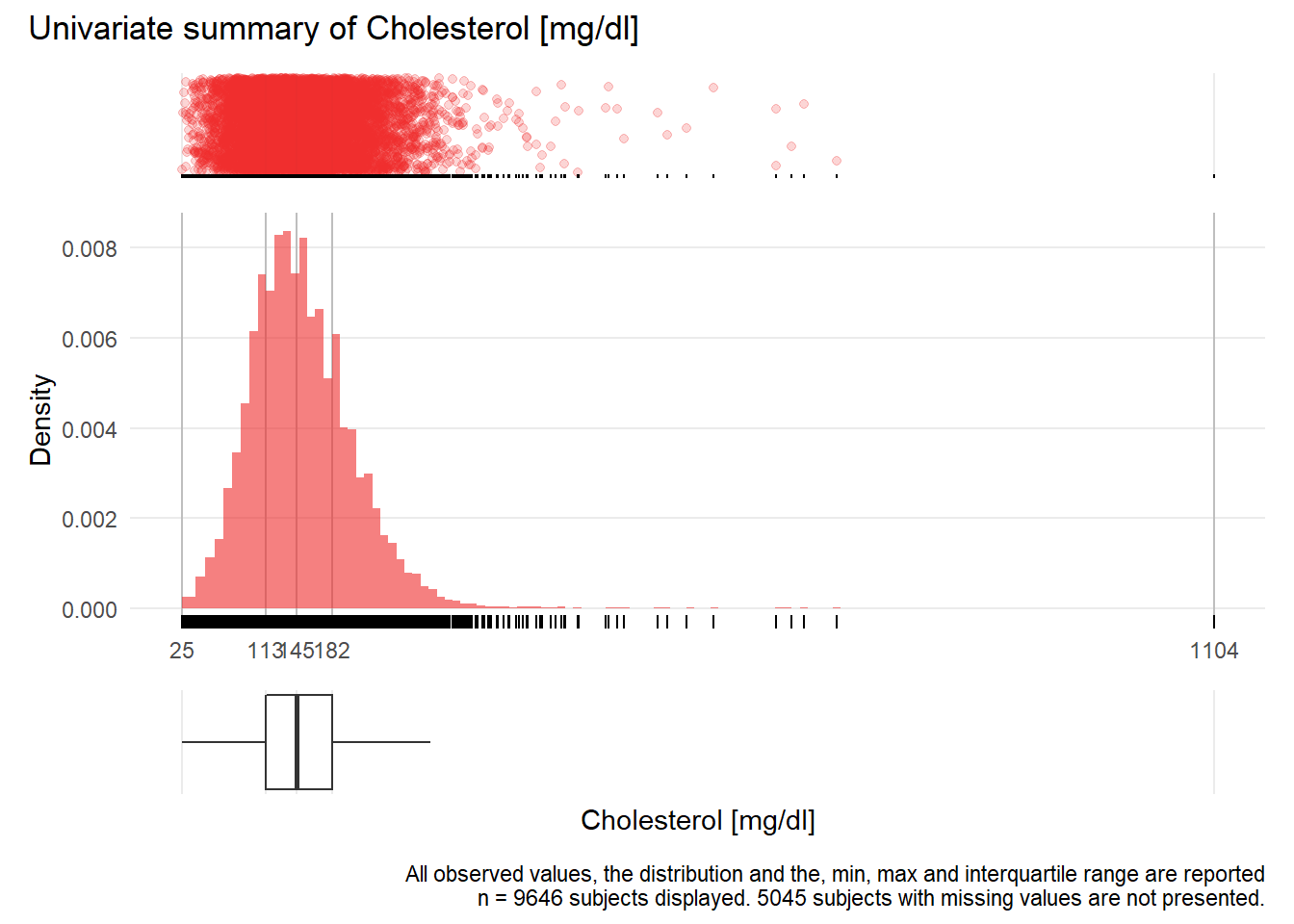
## Warning: Removed 2 rows containing missing values (geom_point).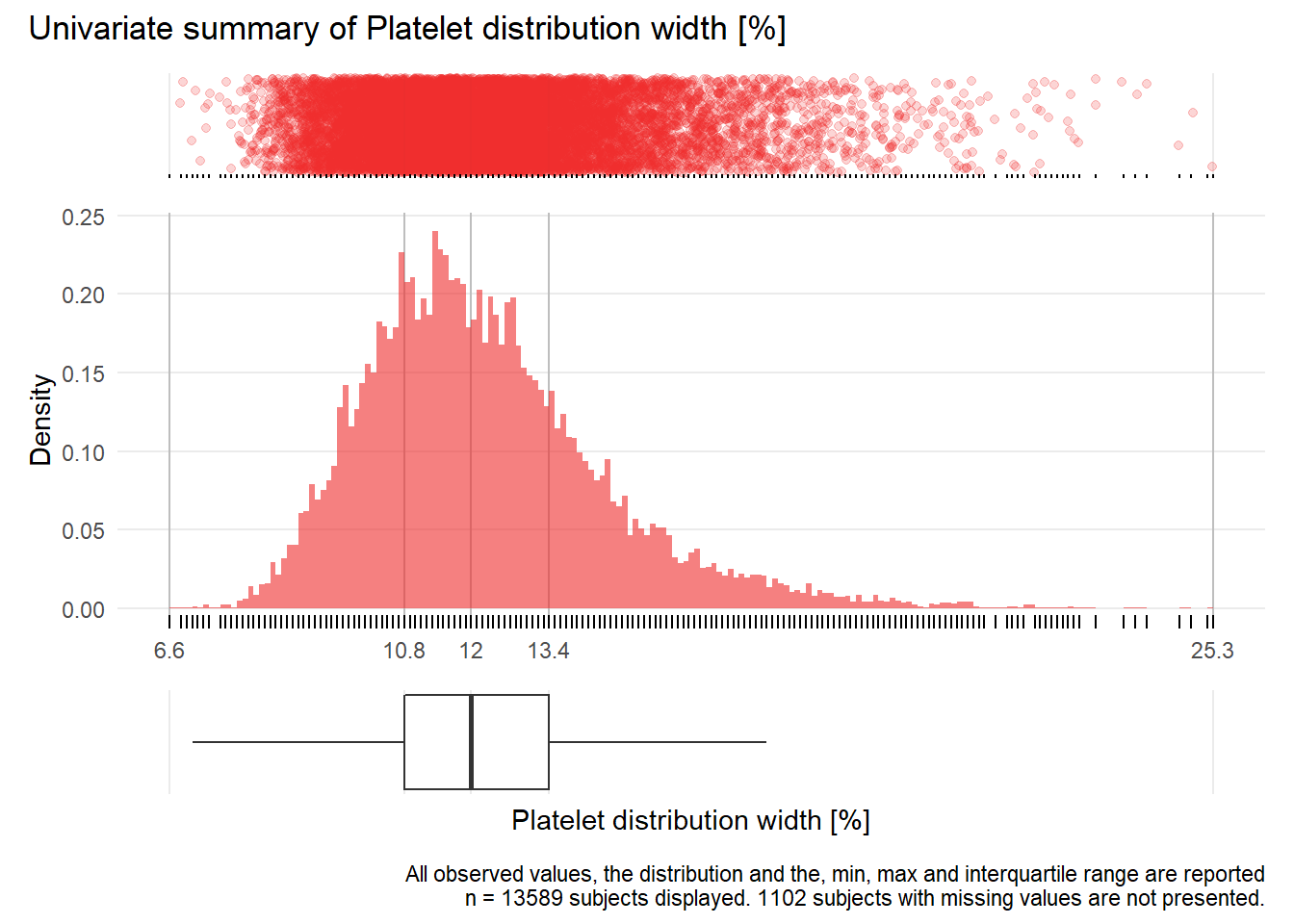
## Warning: Removed 3 rows containing missing values (geom_point).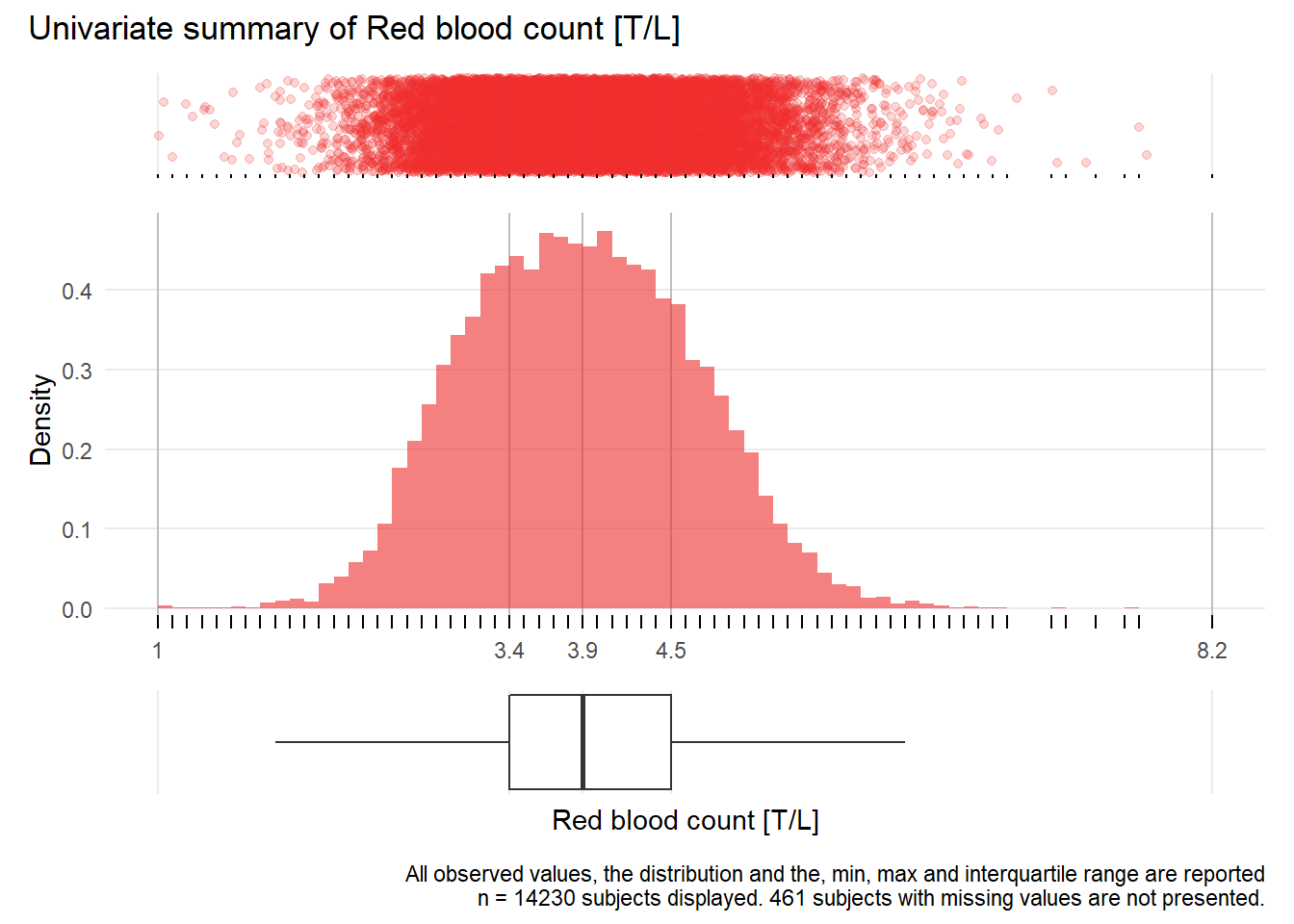
17.3.4 Comparison of univariate distributions with and without pseudo-log transformation
The comparison is only shown for variables where a transformation is suggested.
for(j in 1:length(unique.variables)){
# print(ida_plot_univar_orig_vs_trans(b_bact, unique.variables[j], sigma=res[j], n_bars=100))
if(!is.na(res[j])){
print(ida_plot_univar_orig_vs_trans(b_bact, unique.variables[j], sigma=res[j], n_bars=100))
}
}## Warning: Removed 5 rows containing missing values (geom_point).## Warning: Removed 91 rows containing missing values (geom_point).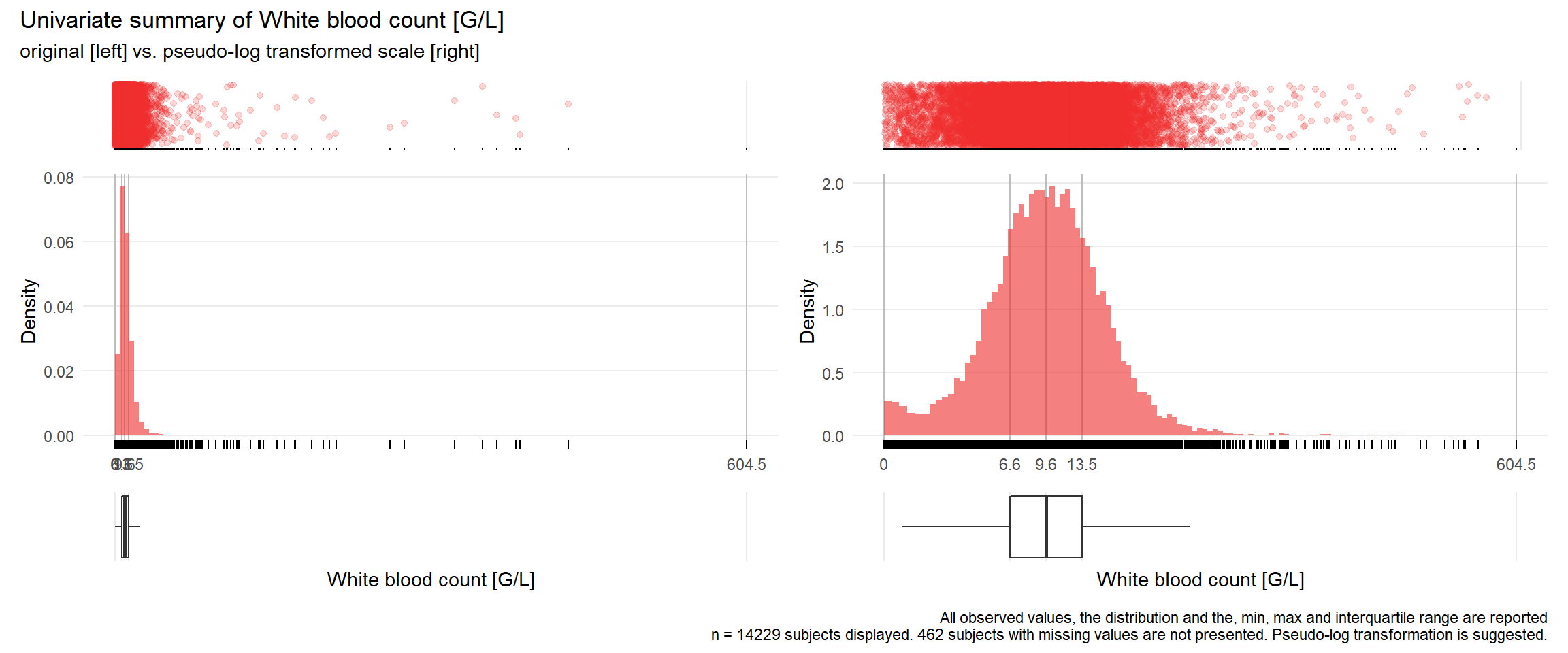
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 8 rows containing missing values (geom_point).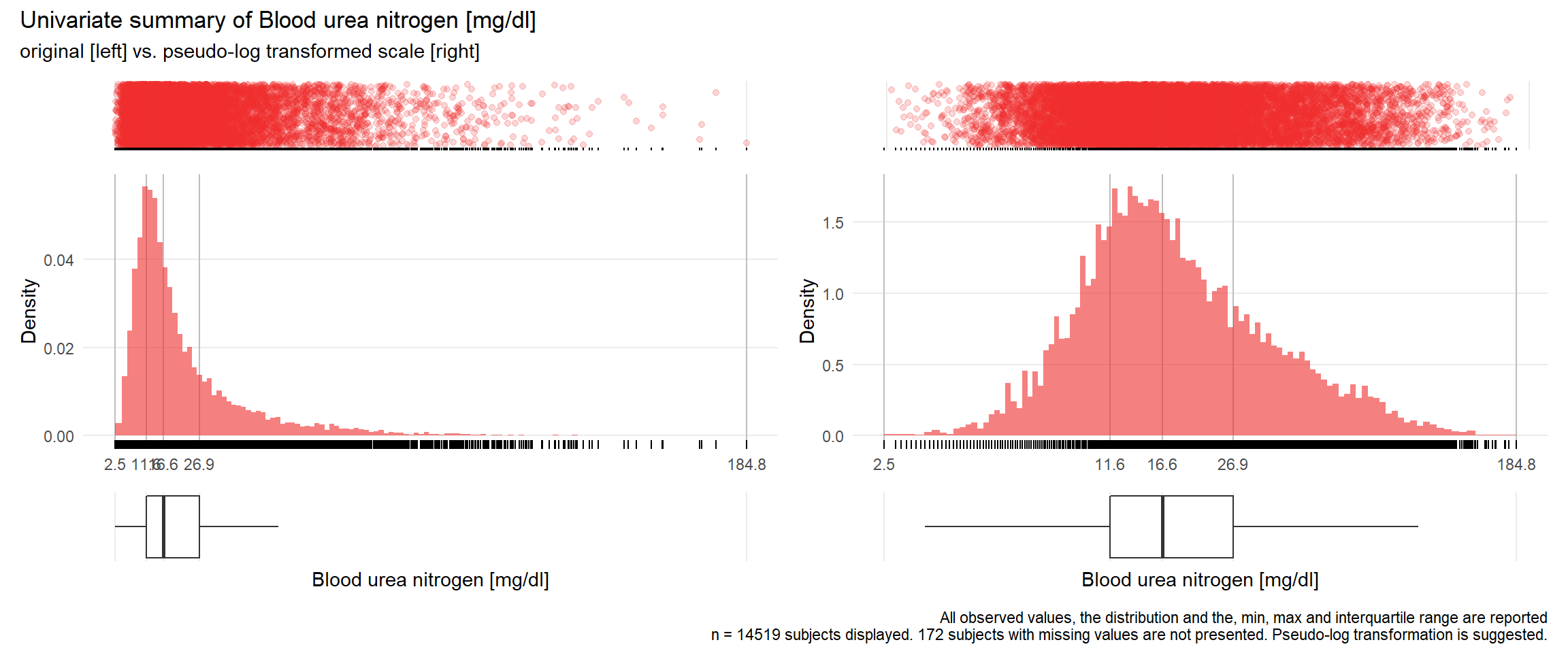
## Warning: Removed 6 rows containing missing values (geom_point).## Warning: Removed 2 rows containing missing values (geom_point).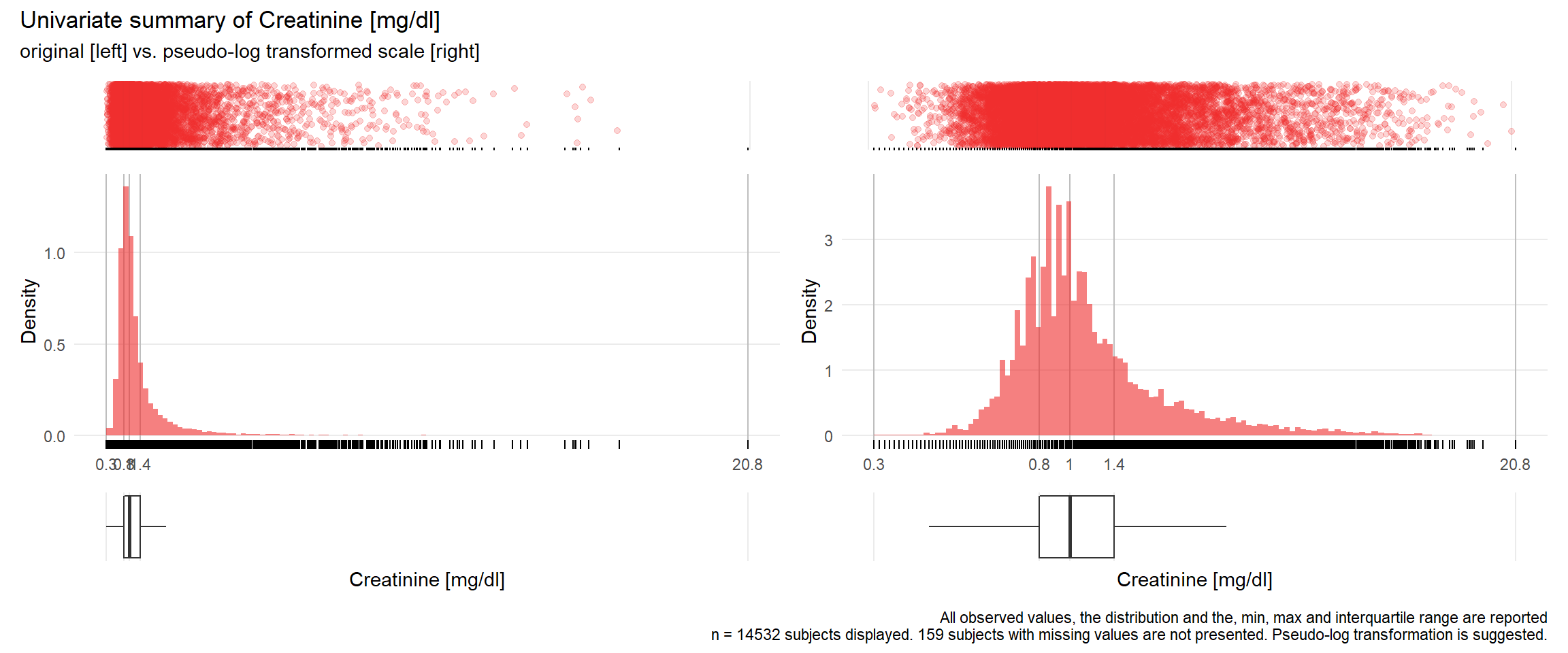
## Warning: Removed 55 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).## Warning: Removed 188 rows containing missing values (geom_point).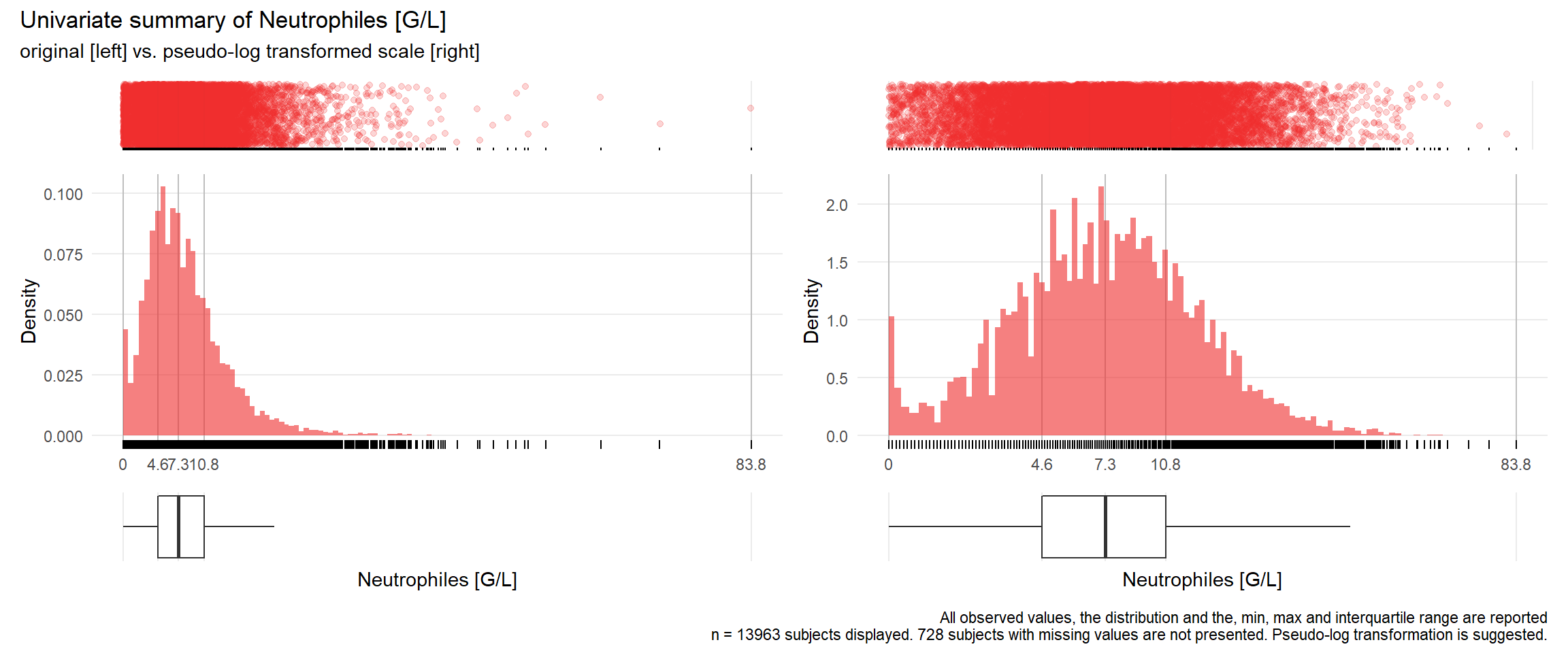
## Warning: Removed 3437 rows containing missing values (geom_point).## Warning: Removed 3417 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).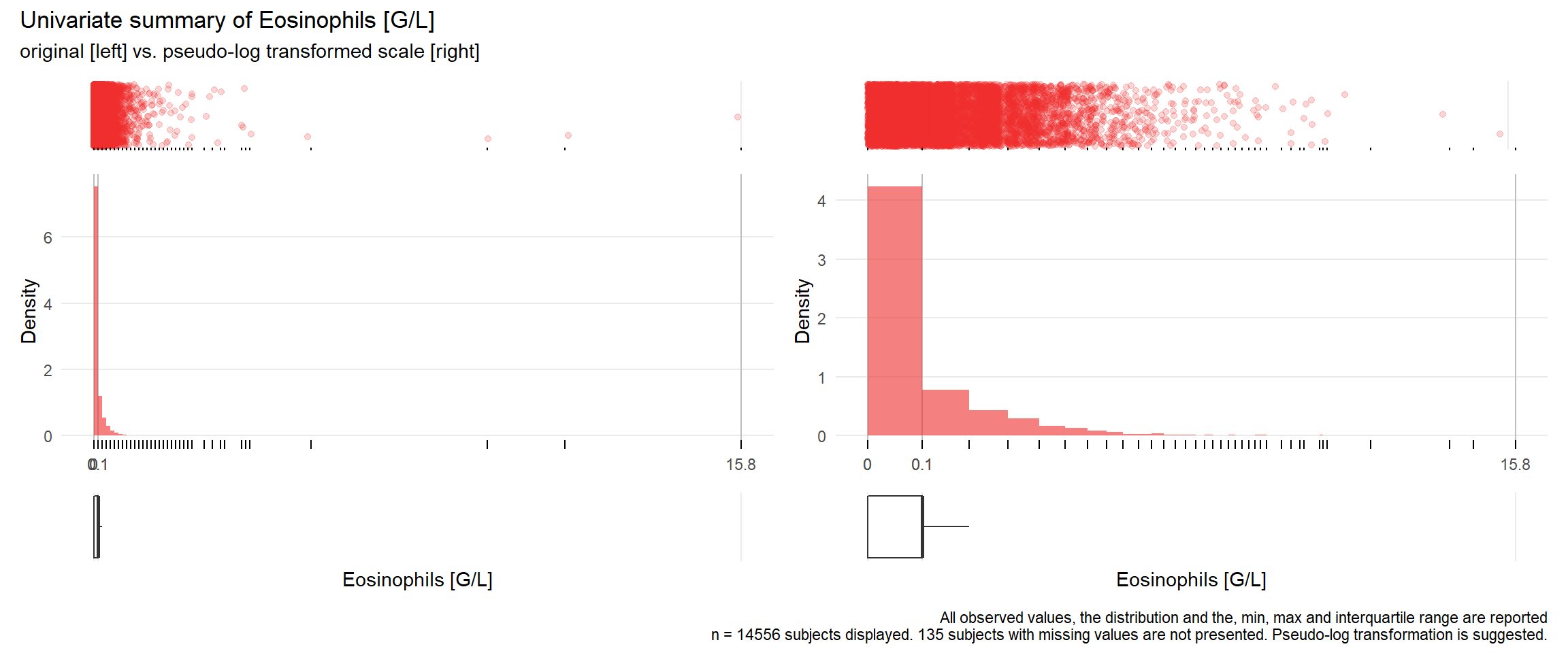
## Warning: Removed 6396 rows containing missing values (geom_point).## Warning: Removed 6306 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).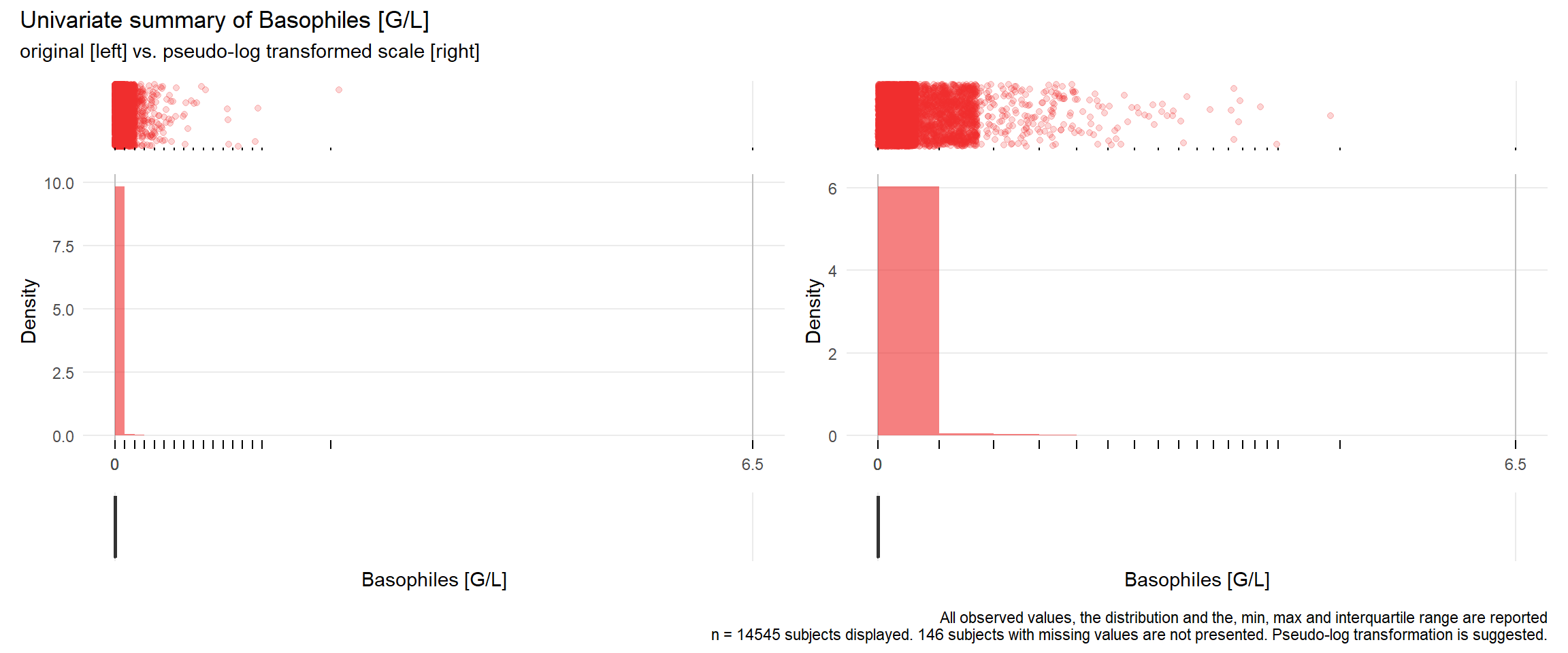
## Warning: Removed 57 rows containing missing values (geom_point).## Warning: Removed 57 rows containing missing values (geom_point).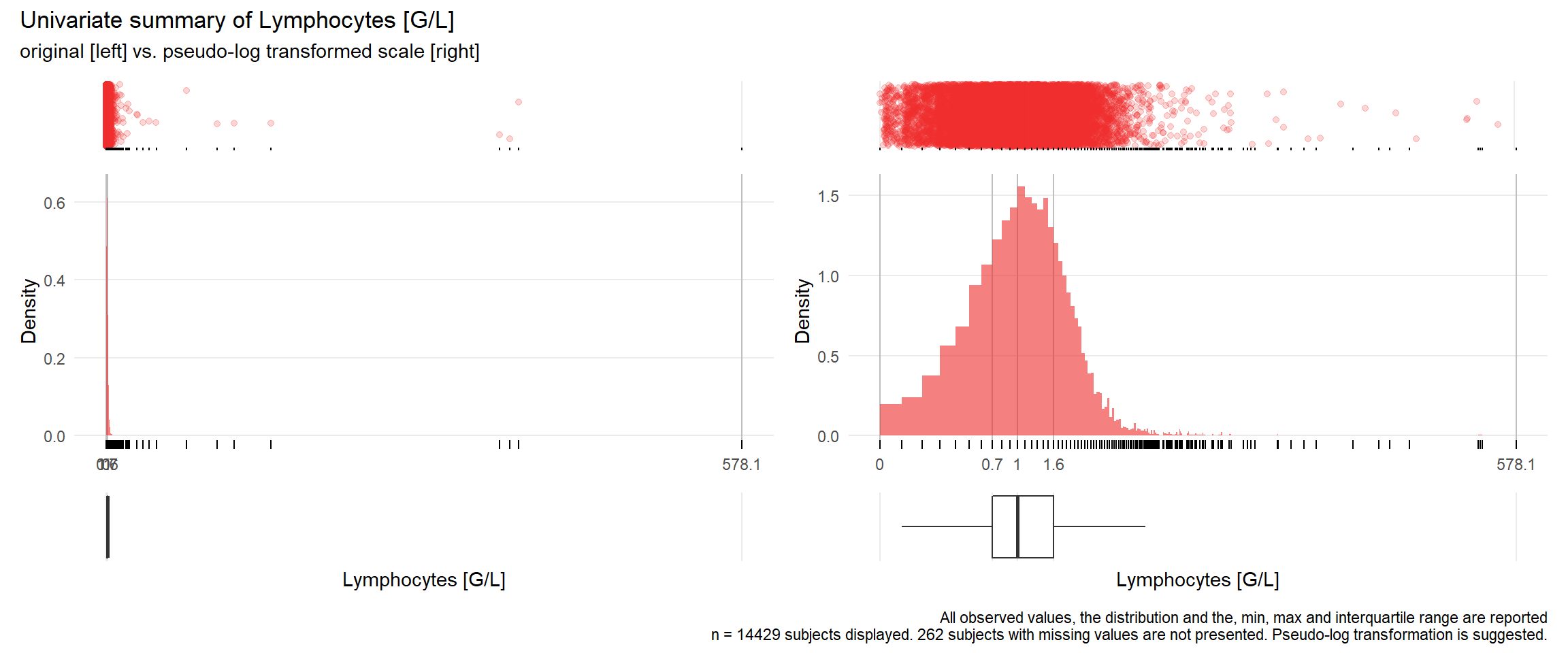
## Warning: Removed 188 rows containing missing values (geom_point).## Warning: Removed 222 rows containing missing values (geom_point).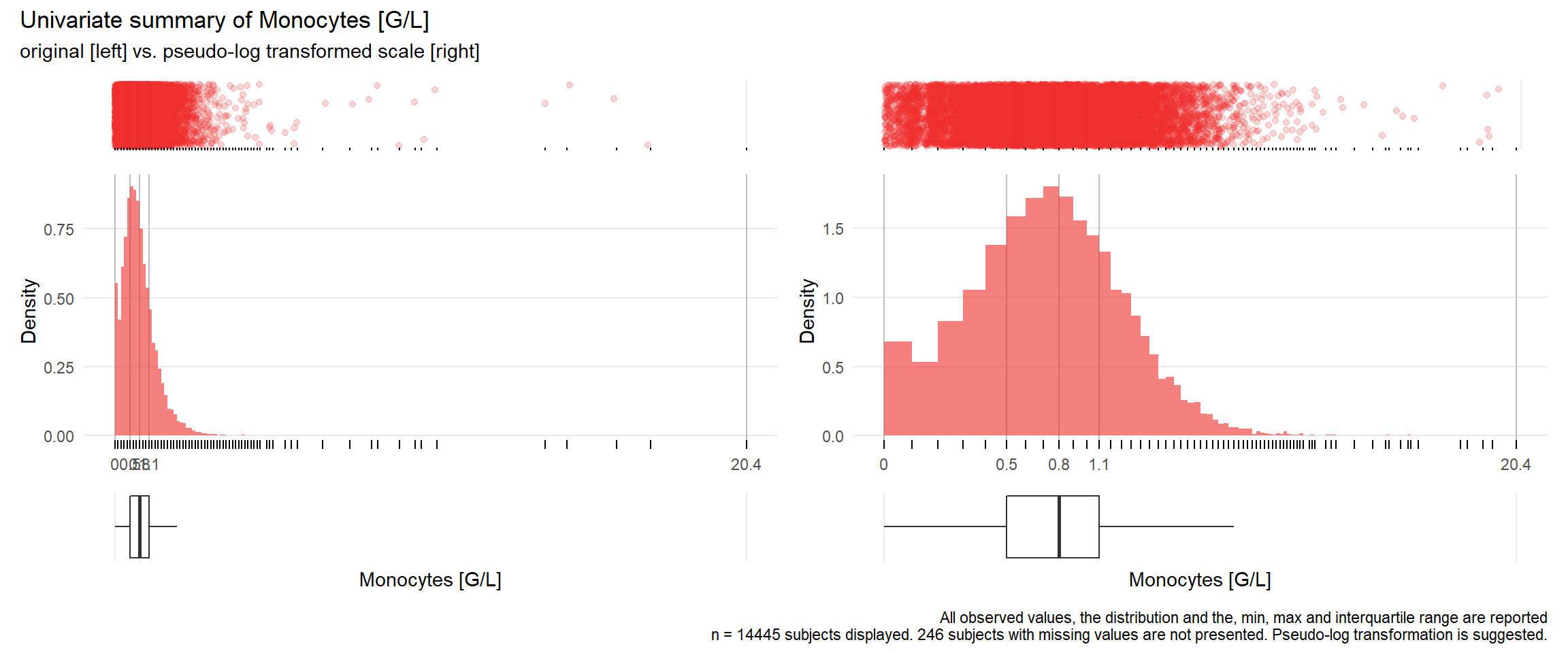
## Warning: Removed 3361 rows containing missing values (geom_point).## Warning: Removed 3381 rows containing missing values (geom_point).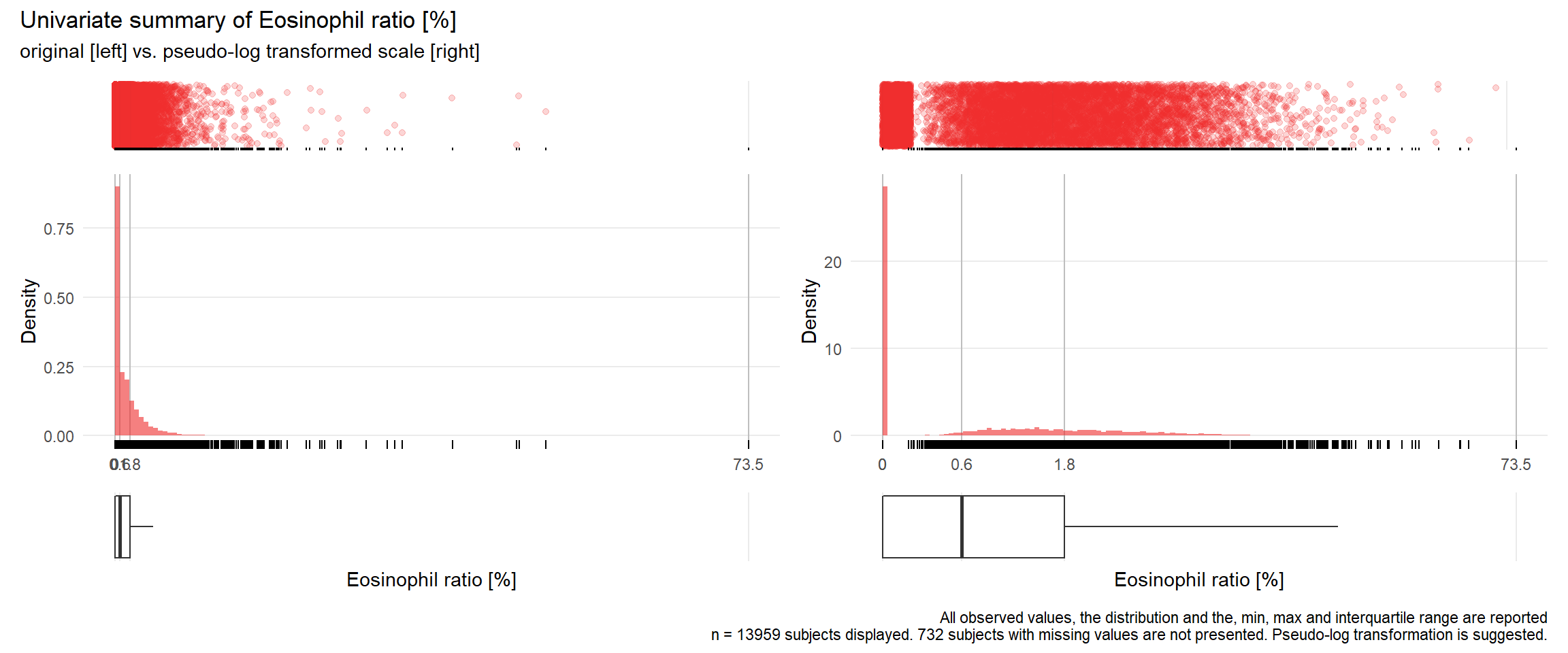
## Warning: Removed 5993 rows containing missing values (geom_point).## Warning: Removed 6175 rows containing missing values (geom_point).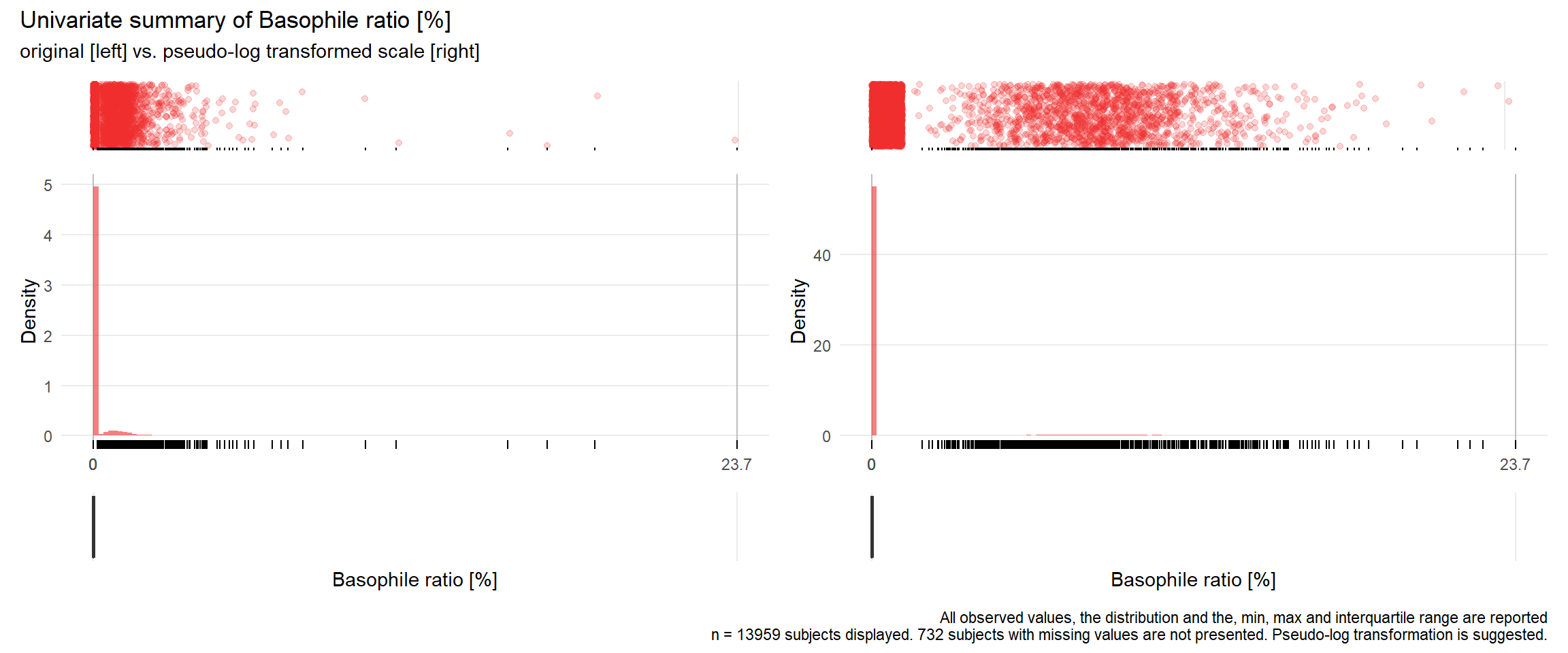
## Warning: Removed 76 rows containing missing values (geom_point).## Warning: Removed 87 rows containing missing values (geom_point).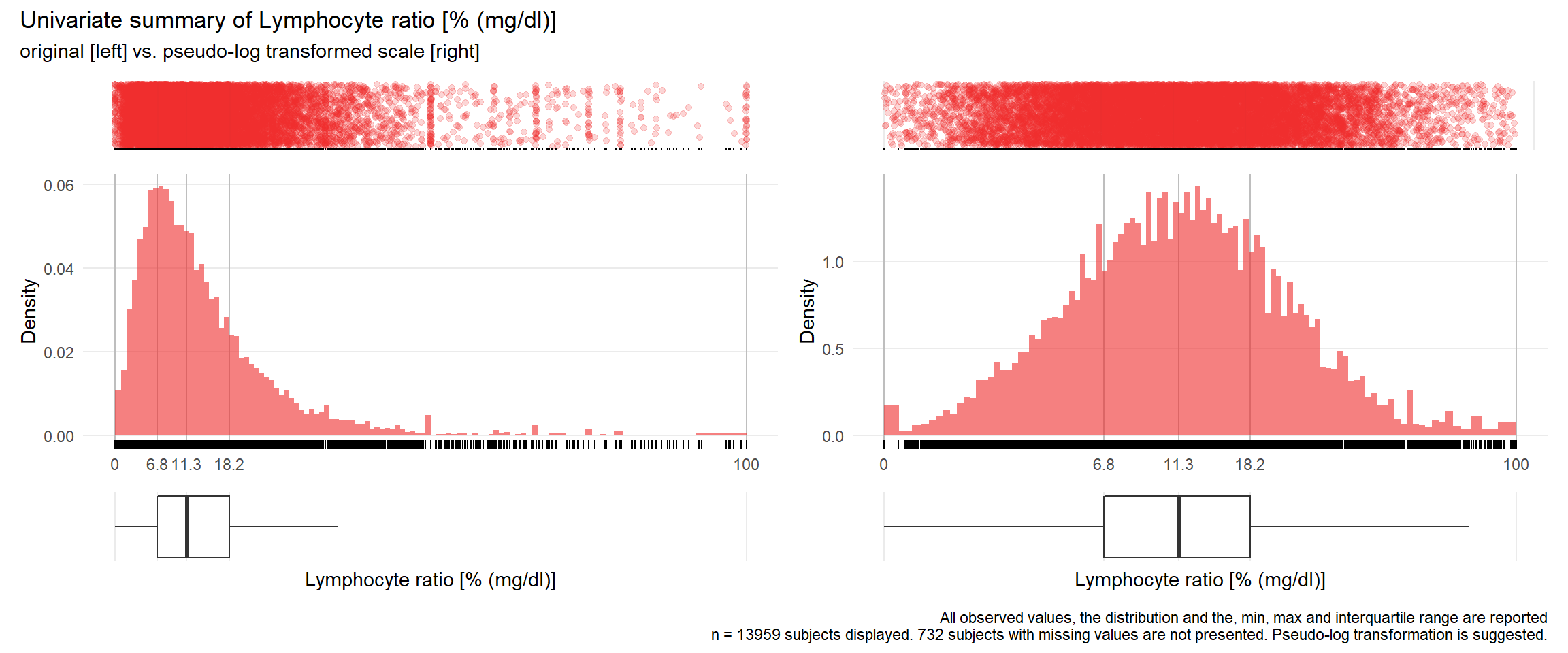
## Warning: Removed 193 rows containing missing values (geom_point).## Warning: Removed 204 rows containing missing values (geom_point).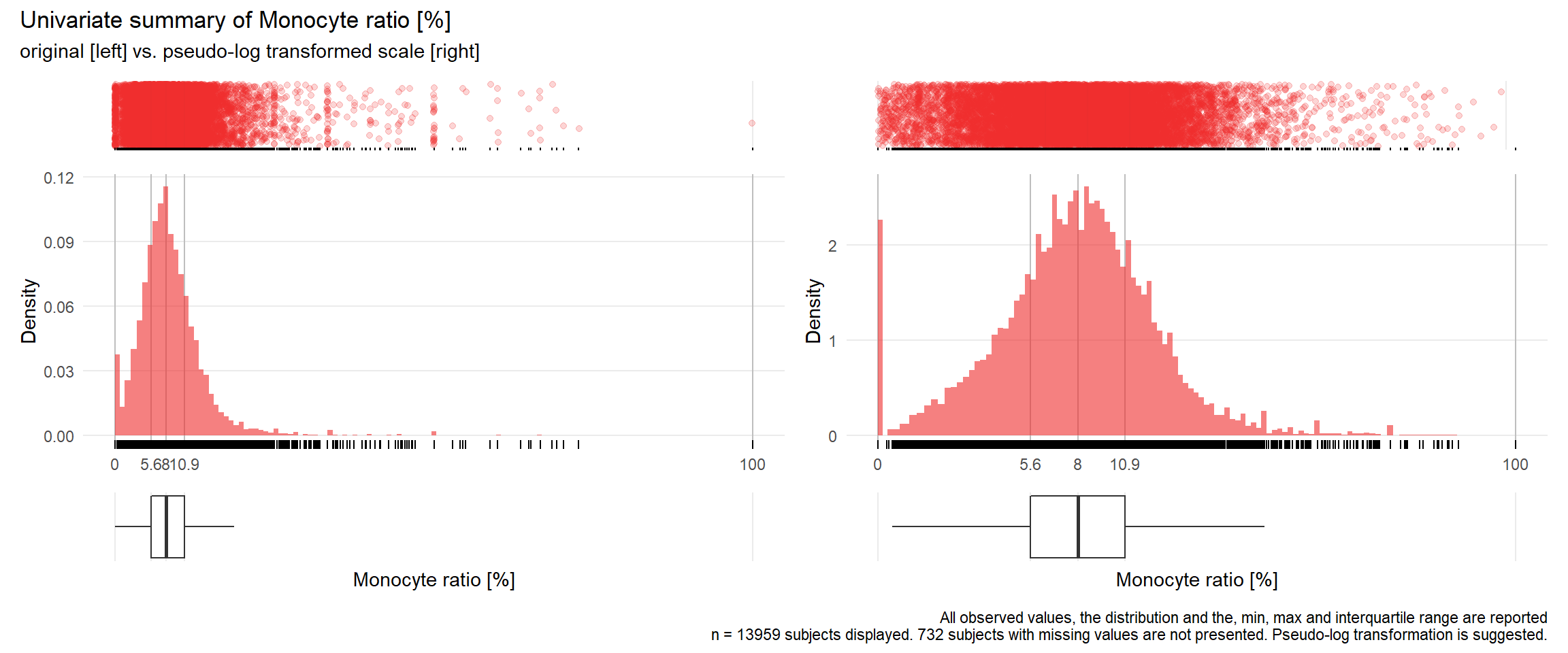
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 3 rows containing missing values (geom_point).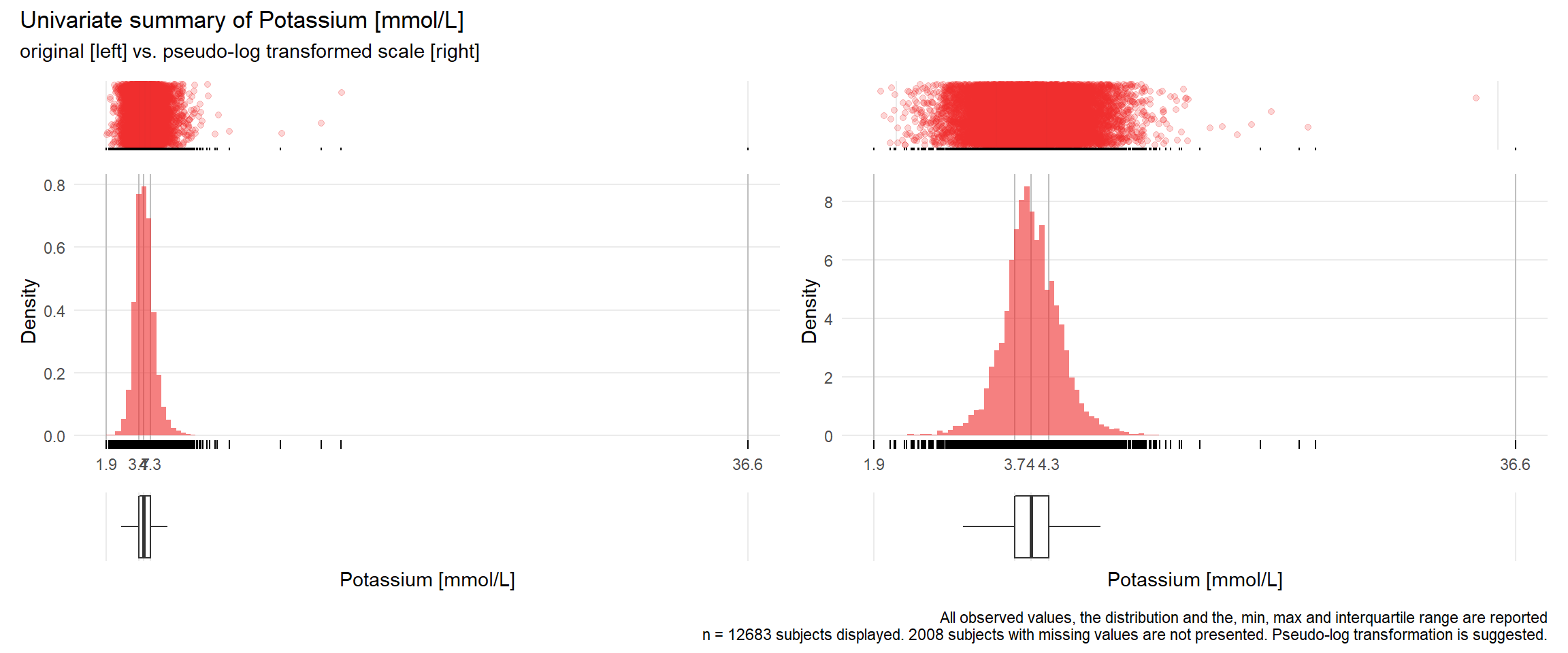
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_point).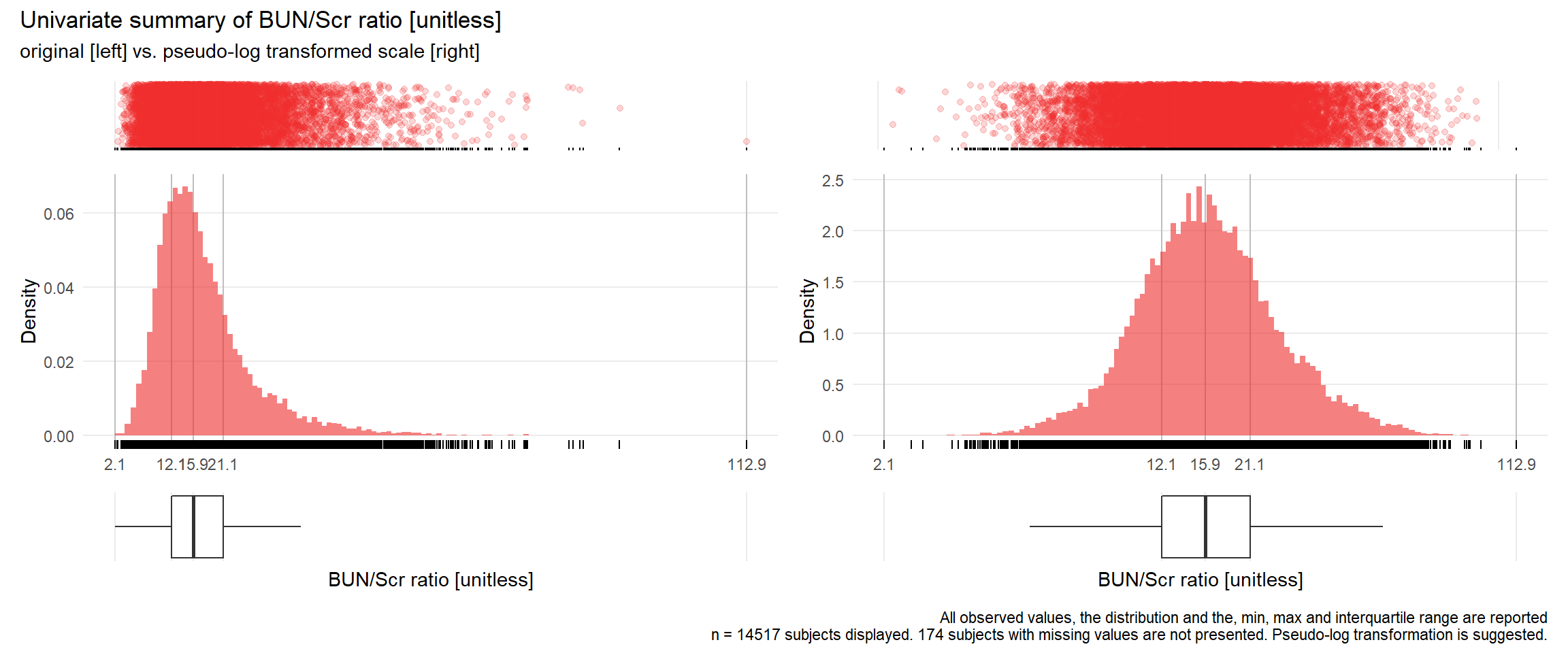
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).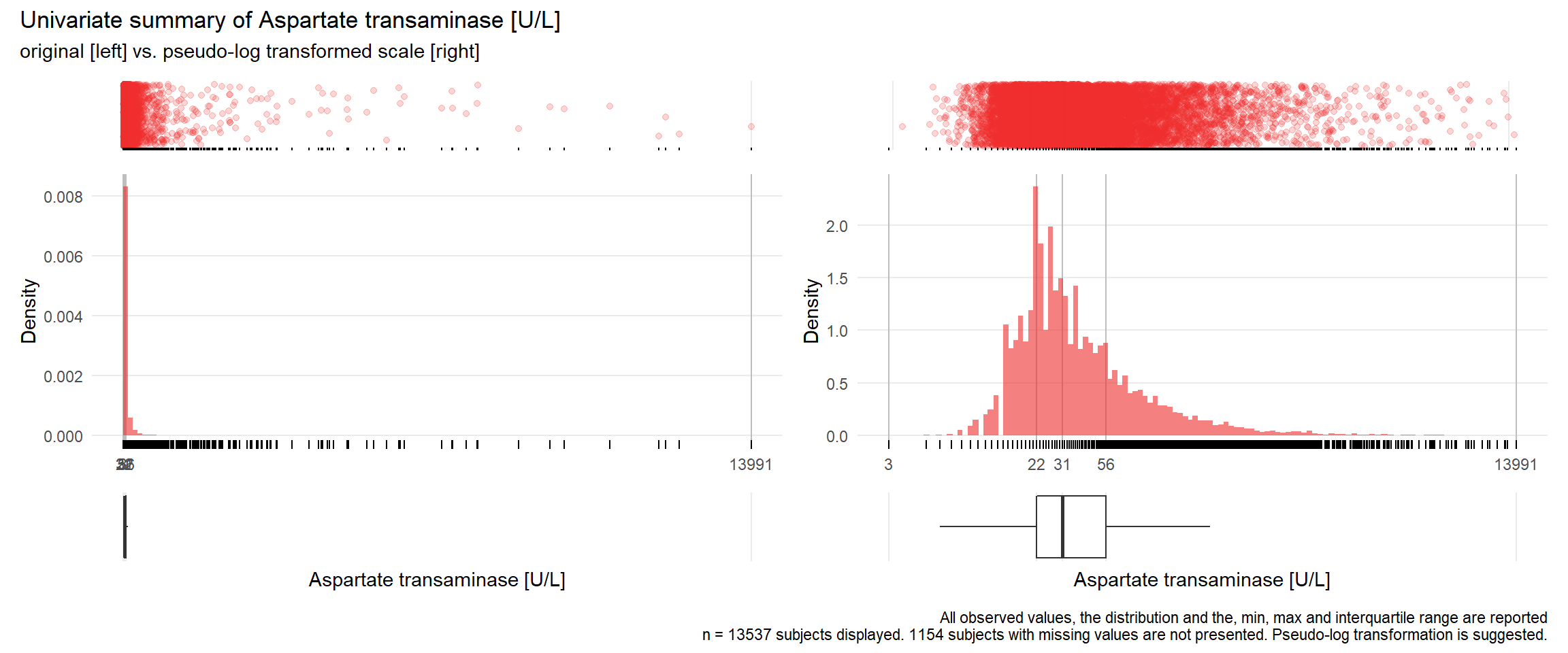
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_point).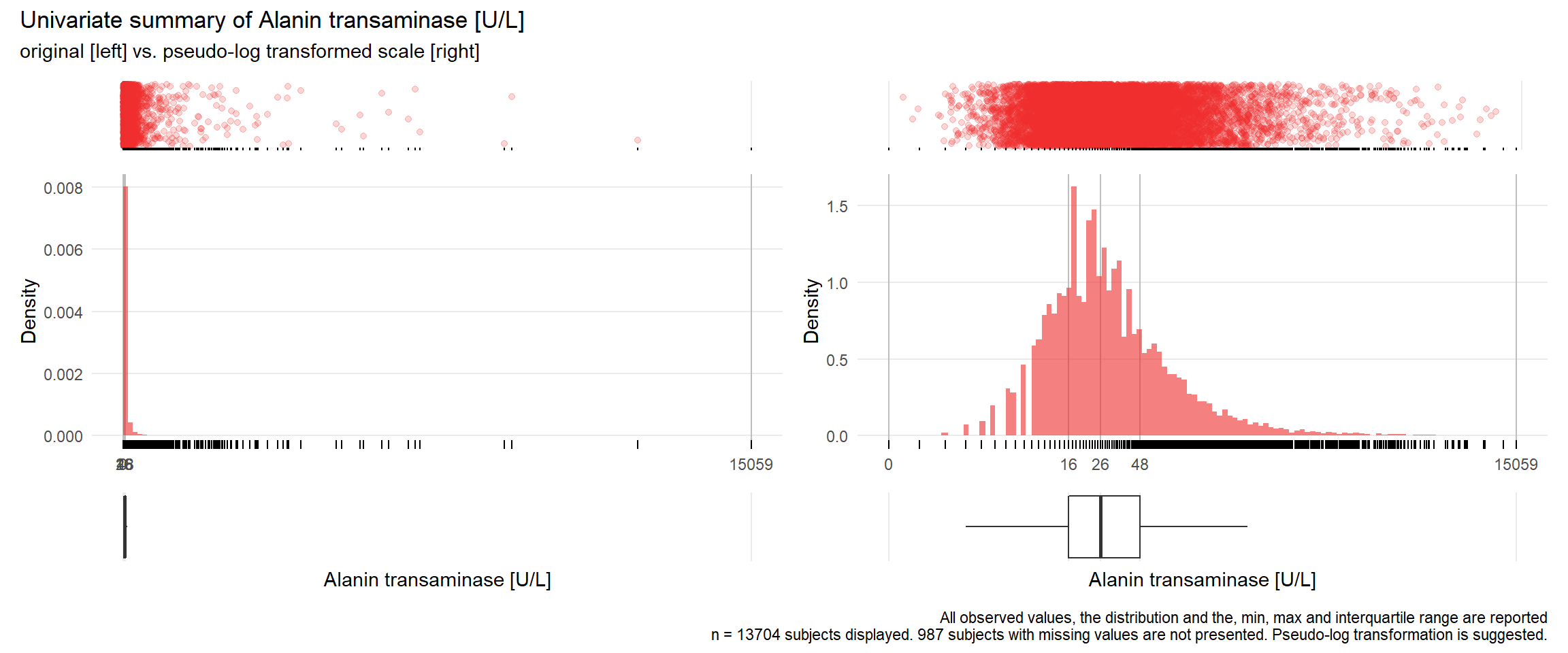
## Warning: Removed 2 rows containing missing values (geom_point).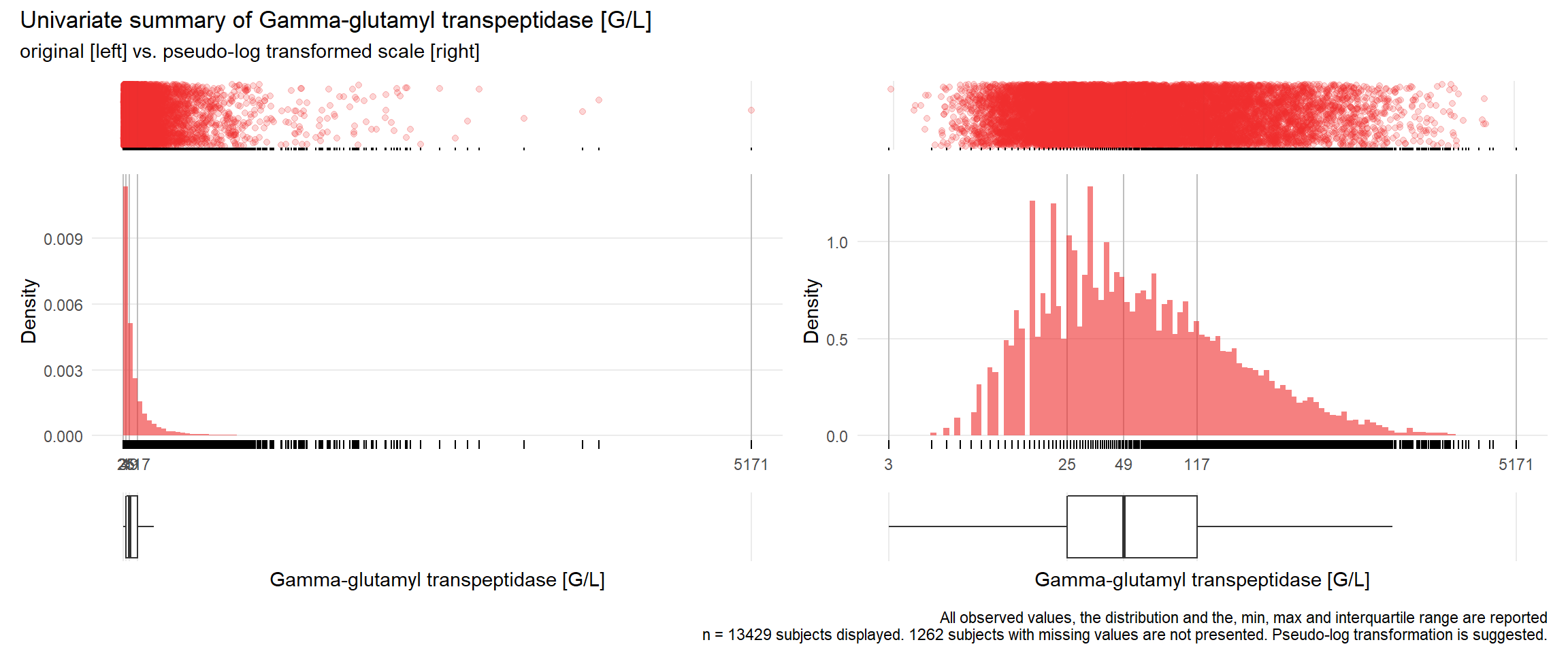
## Warning: Removed 6 rows containing missing values (geom_point).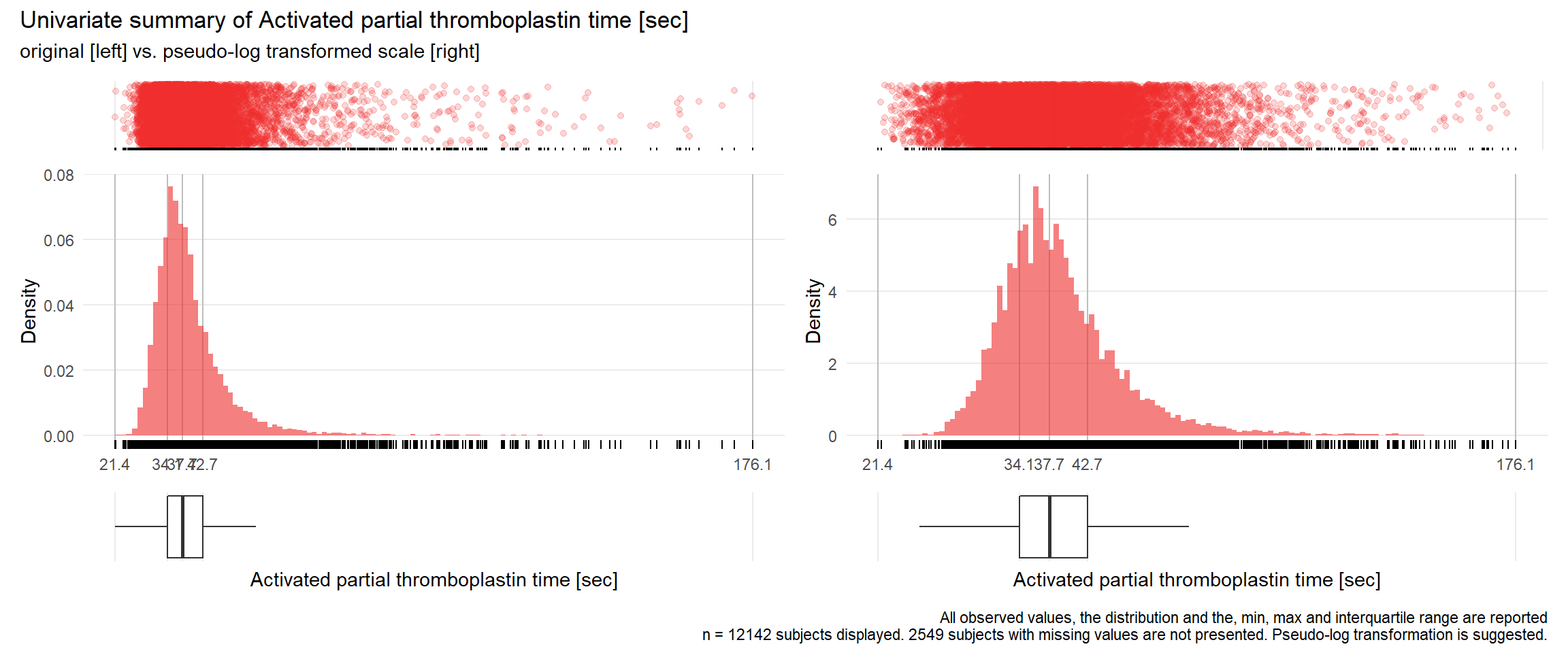
## Warning: Removed 32 rows containing missing values (geom_point).## Warning: Removed 22 rows containing missing values (geom_point).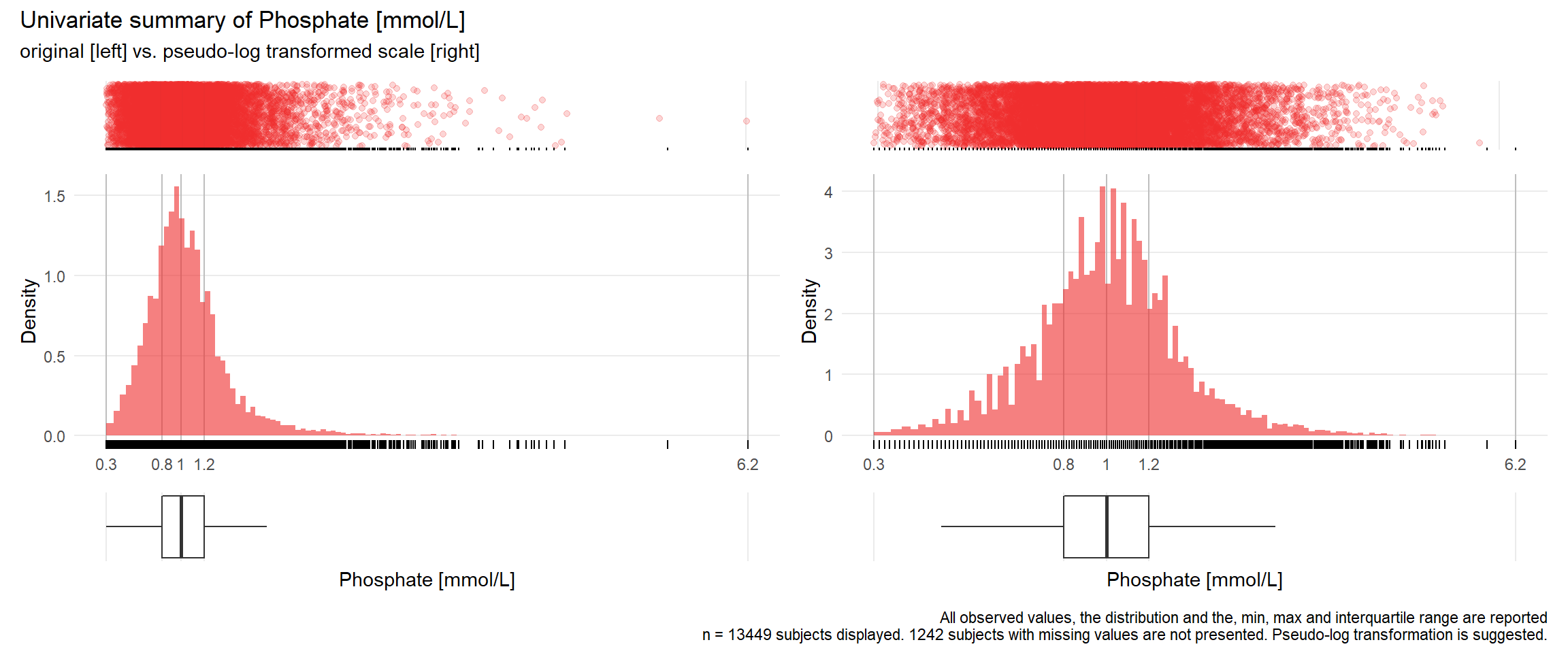
## Warning: Removed 7 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).## Warning: Removed 5 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).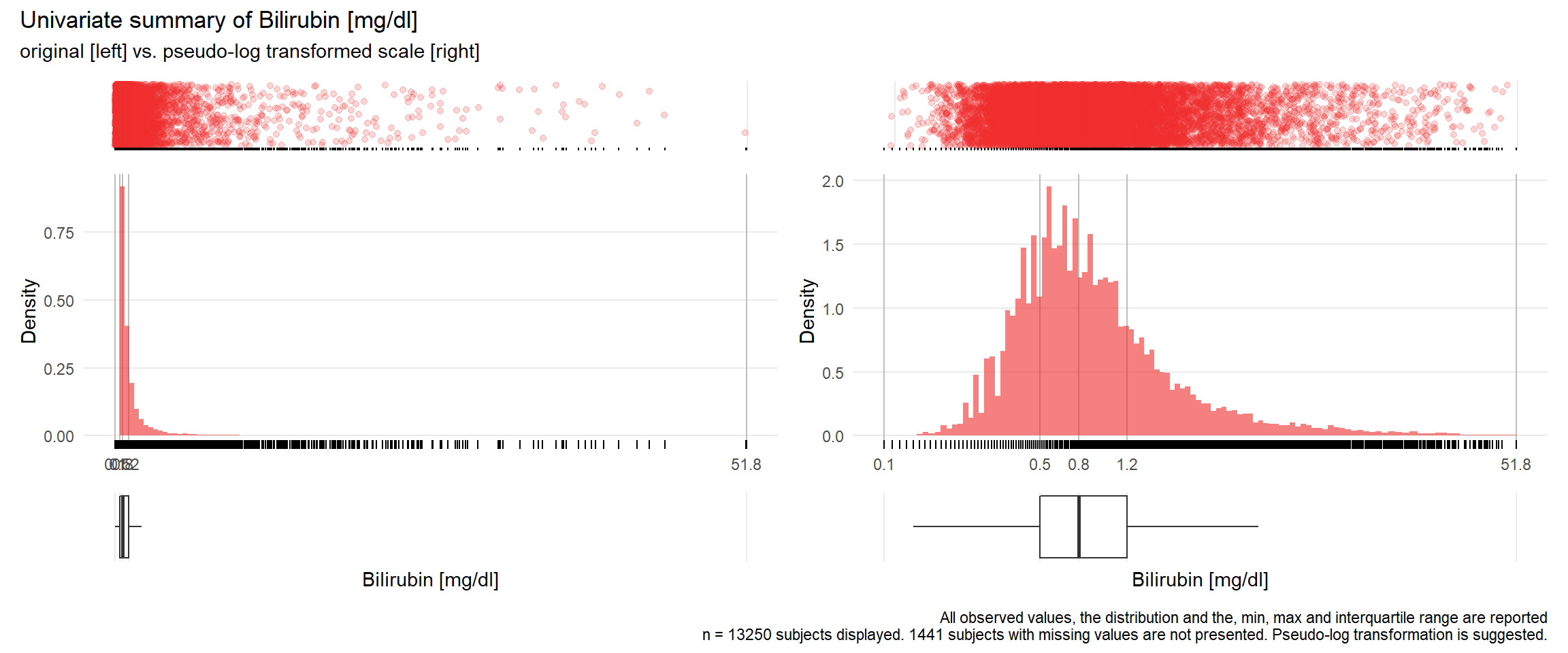
## Warning: Removed 2 rows containing missing values (geom_point).## Warning: Removed 7 rows containing missing values (geom_point).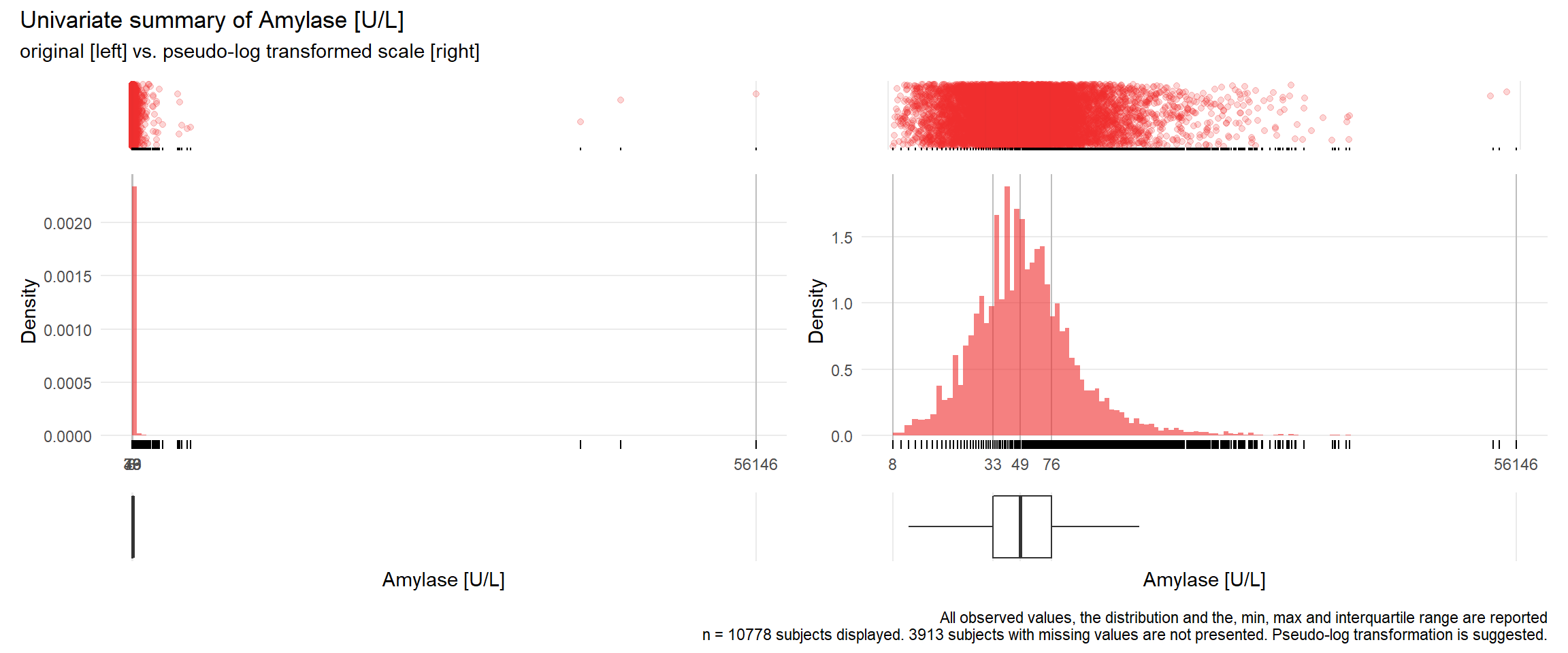
## Warning: Removed 3 rows containing missing values (geom_point).## Warning: Removed 3 rows containing missing values (geom_point).## Warning: Removed 1 rows containing missing values (geom_bar).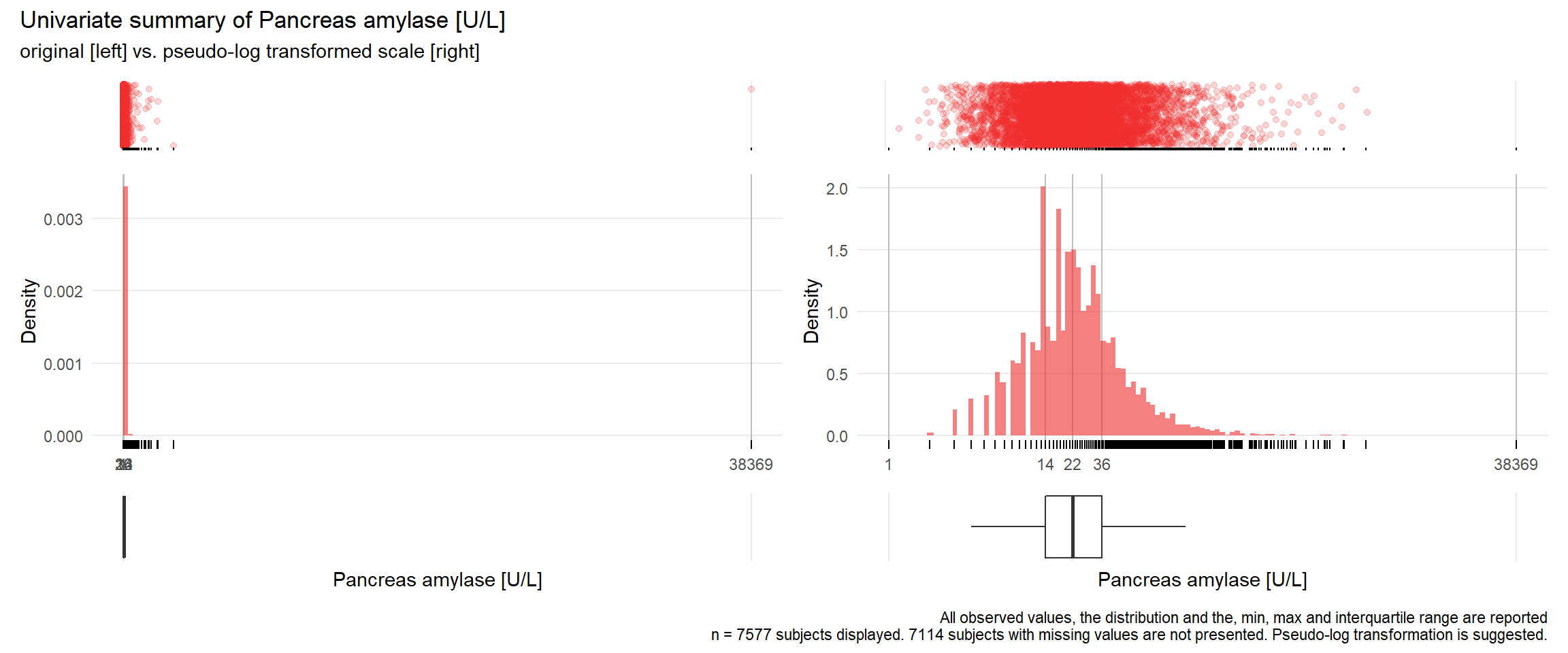
## Warning: Removed 1 rows containing missing values (geom_point).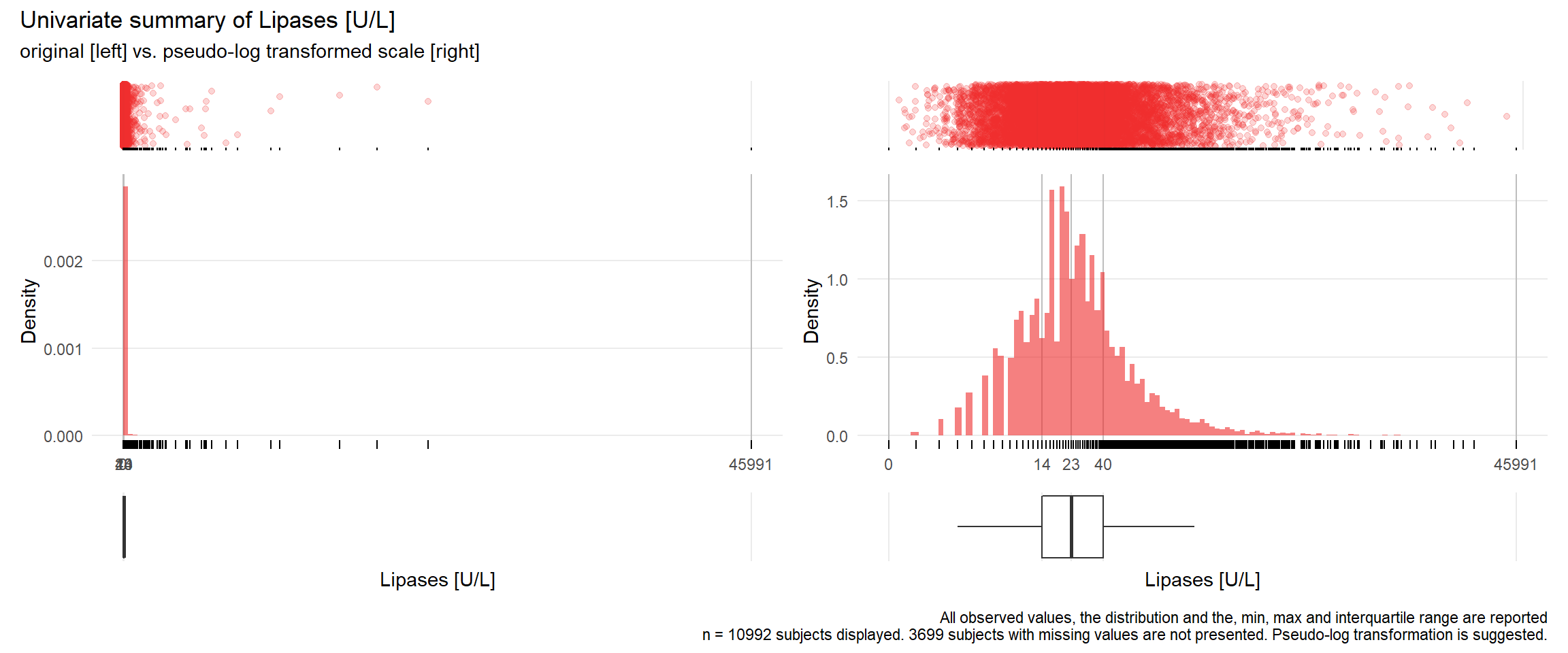
## Warning: Removed 1 rows containing missing values (geom_point).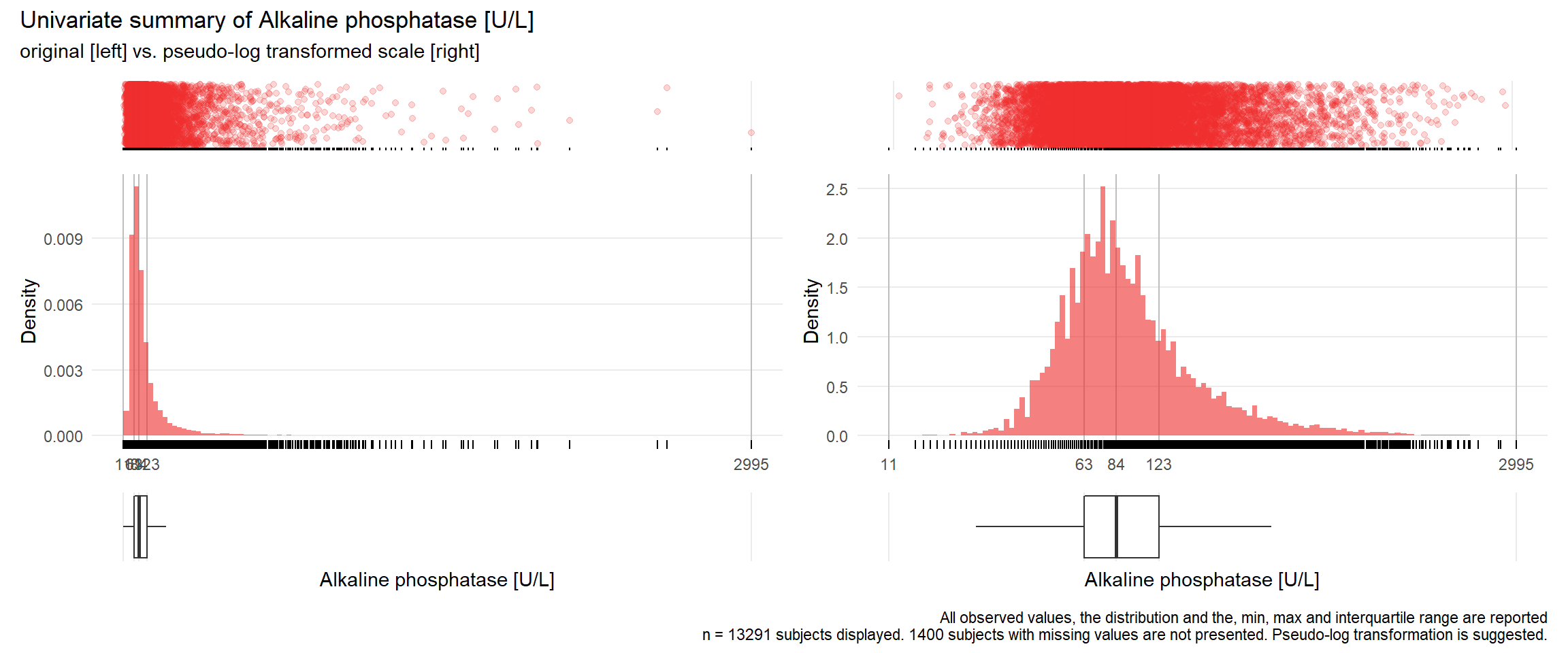
## Warning: Removed 2 rows containing missing values (geom_point).
## Removed 1 rows containing missing values (geom_bar).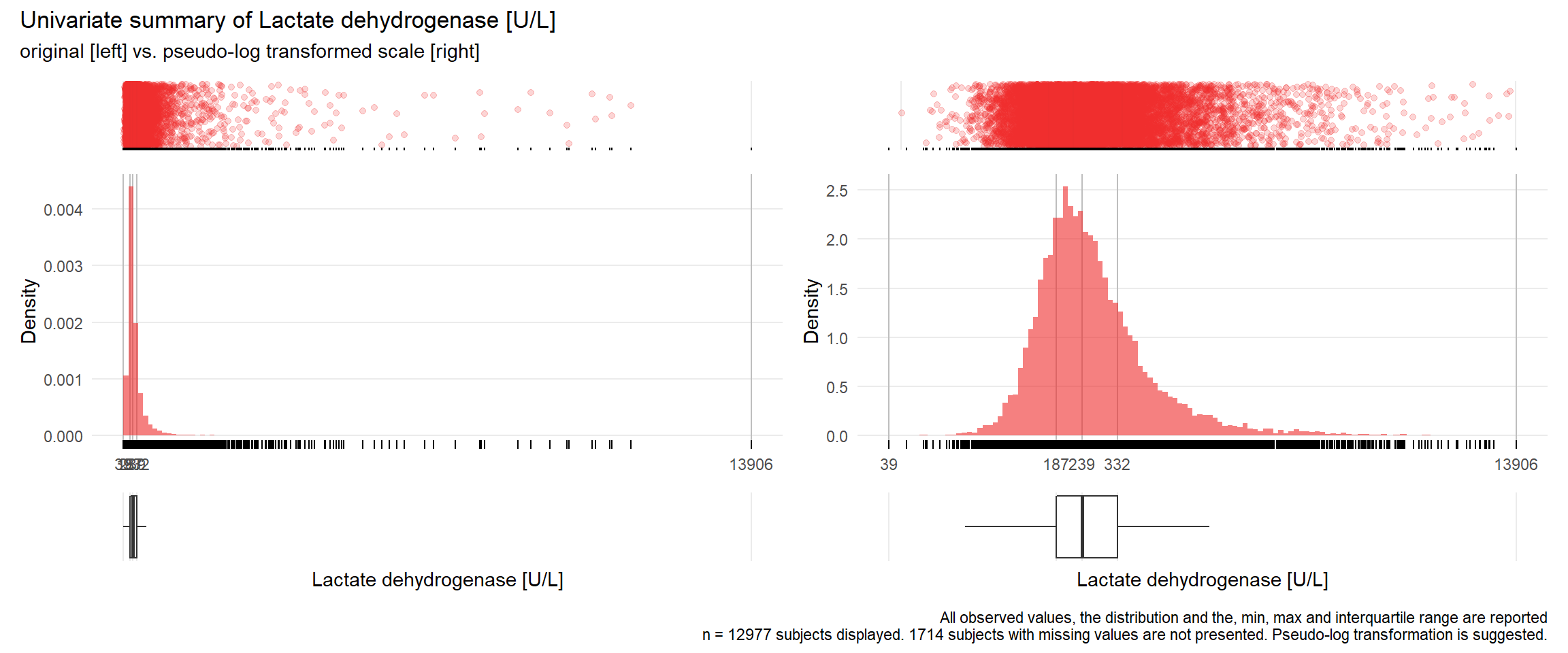
## Warning: Removed 1 rows containing missing values (geom_point).## Warning: Removed 5 rows containing missing values (geom_point).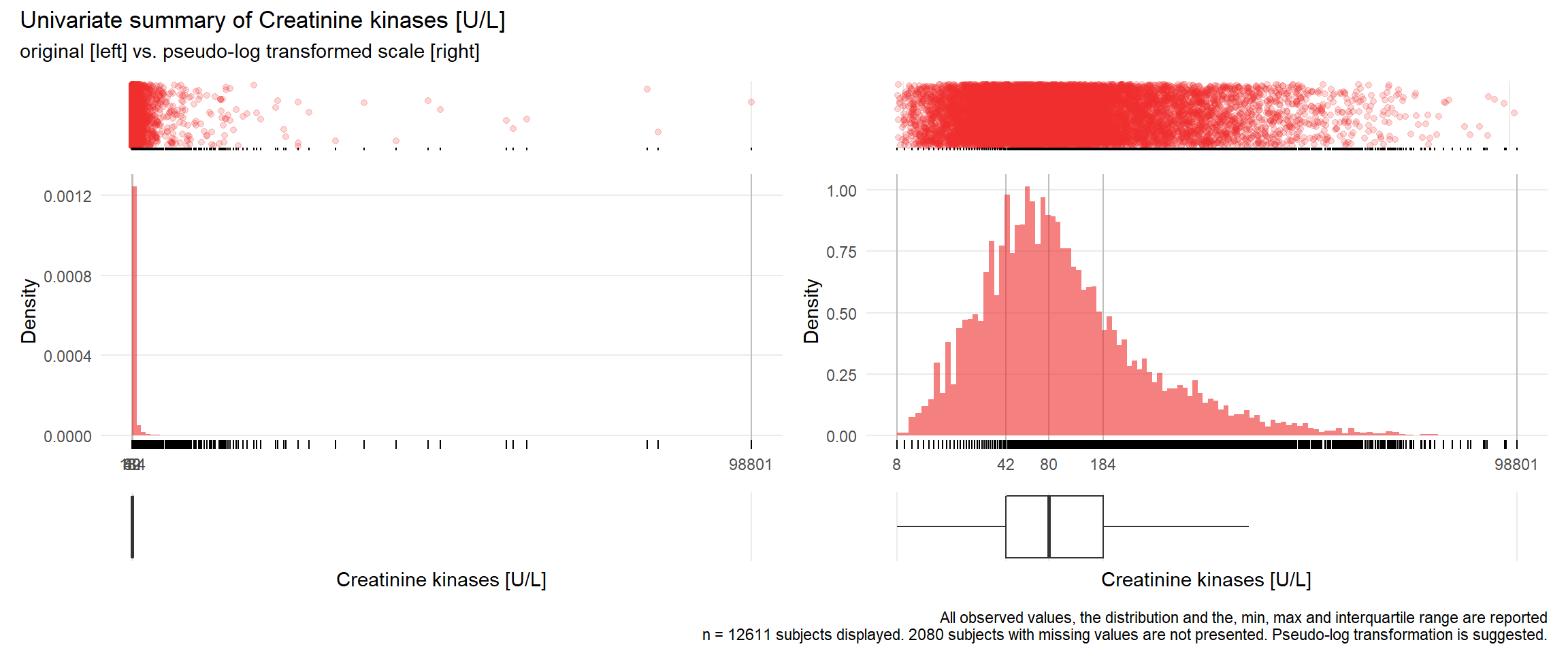
## Warning: Removed 2 rows containing missing values (geom_point).## Warning: Removed 2 rows containing missing values (geom_point).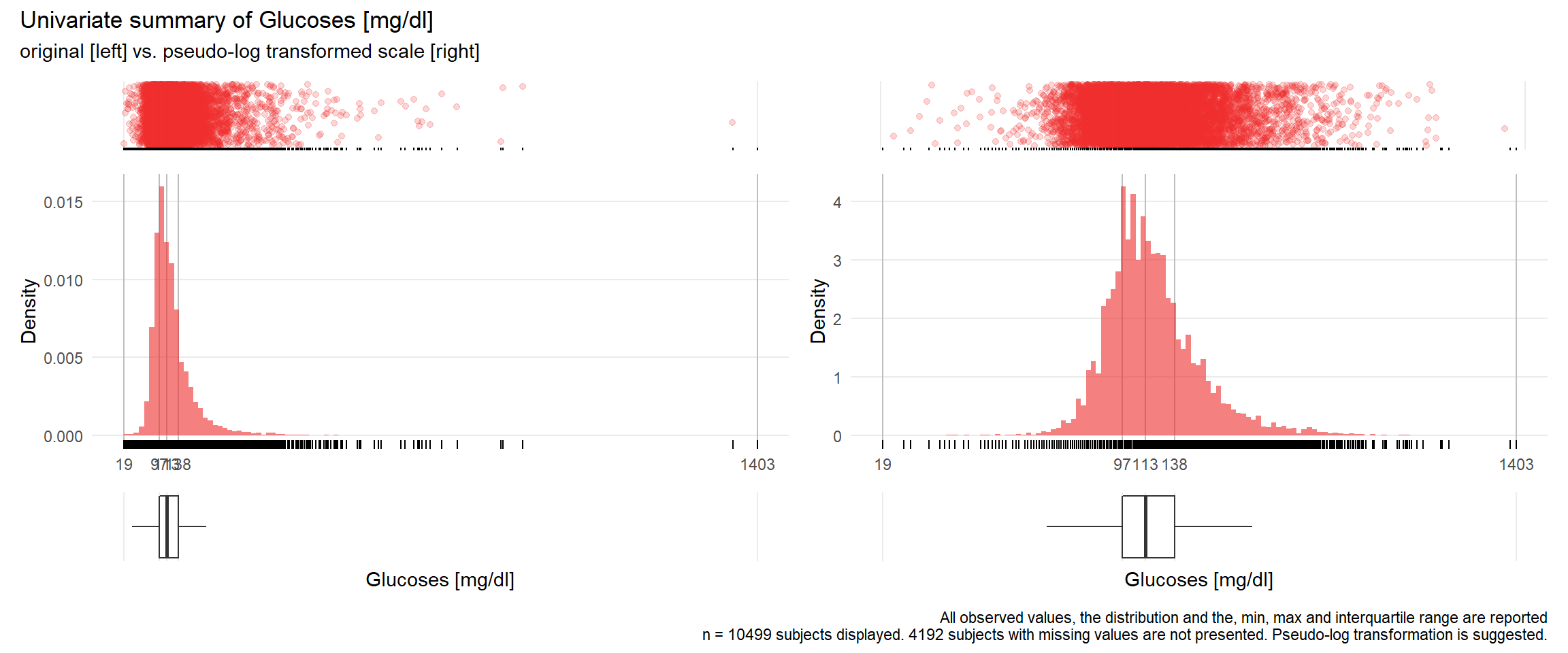
## Warning: Removed 2 rows containing missing values (geom_point).## Warning: Removed 3 rows containing missing values (geom_point).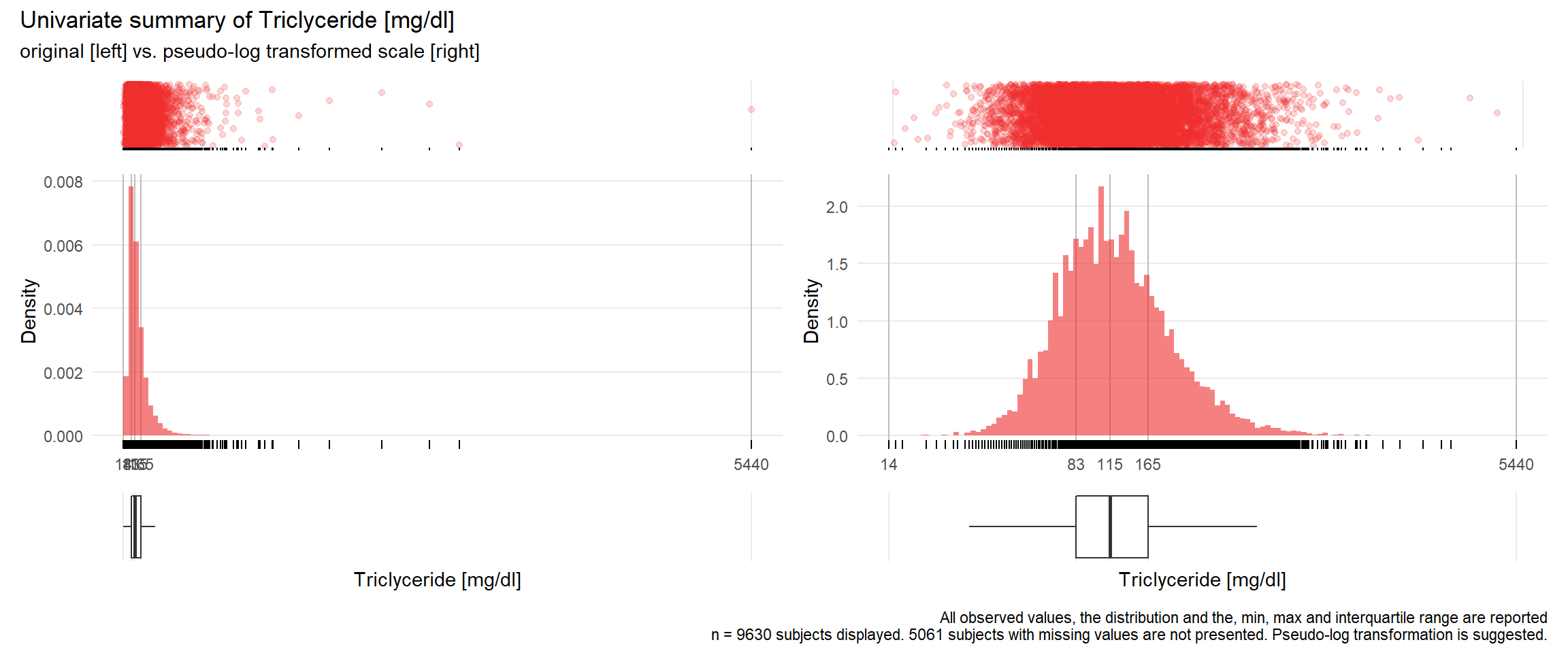
17.4 Section session info
## R version 4.1.3 (2022-03-10)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_Austria.1252 LC_CTYPE=English_Austria.1252
## [3] LC_MONETARY=English_Austria.1252 LC_NUMERIC=C
## [5] LC_TIME=English_Austria.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] Hmisc_4.6-0 Formula_1.2-4 survival_3.2-13 lattice_0.20-45
## [5] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.8 purrr_0.3.4
## [9] readr_2.1.2 tidyr_1.2.0 tibble_3.1.6 ggplot2_3.3.5
## [13] tidyverse_1.3.1 here_1.0.1
##
## loaded via a namespace (and not attached):
## [1] fs_1.5.2 lubridate_1.8.0 RColorBrewer_1.1-2
## [4] httr_1.4.2 rprojroot_2.0.2 tools_4.1.3
## [7] backports_1.4.1 bslib_0.3.1 utf8_1.2.2
## [10] R6_2.5.1 rpart_4.1.16 DBI_1.1.2
## [13] colorspace_2.0-3 nnet_7.3-17 withr_2.5.0
## [16] tidyselect_1.1.2 gridExtra_2.3 compiler_4.1.3
## [19] cli_3.2.0 rvest_1.0.2 htmlTable_2.4.0
## [22] xml2_1.3.3 labeling_0.4.2 bookdown_0.25
## [25] sass_0.4.1 scales_1.1.1 checkmate_2.0.0
## [28] digest_0.6.29 foreign_0.8-82 rmarkdown_2.13
## [31] base64enc_0.1-3 jpeg_0.1-9 pkgconfig_2.0.3
## [34] htmltools_0.5.2 highr_0.9 dbplyr_2.1.1
## [37] fastmap_1.1.0 htmlwidgets_1.5.4 rlang_1.0.2
## [40] readxl_1.3.1 rstudioapi_0.13 jquerylib_0.1.4
## [43] generics_0.1.2 farver_2.1.0 jsonlite_1.8.0
## [46] magrittr_2.0.2 patchwork_1.1.1 Matrix_1.4-0
## [49] Rcpp_1.0.8.3 munsell_0.5.0 fansi_1.0.3
## [52] lifecycle_1.0.1 stringi_1.7.6 yaml_2.3.5
## [55] grid_4.1.3 crayon_1.5.1 haven_2.4.3
## [58] splines_4.1.3 hms_1.1.1 knitr_1.38
## [61] pillar_1.7.0 reprex_2.0.1 glue_1.6.2
## [64] evaluate_0.15 latticeExtra_0.6-29 data.table_1.14.2
## [67] modelr_0.1.8 png_0.1-7 vctrs_0.3.8
## [70] tzdb_0.2.0 cellranger_1.1.0 gtable_0.3.0
## [73] assertthat_0.2.1 xfun_0.30 broom_0.7.12
## [76] cluster_2.1.2 ellipsis_0.3.2