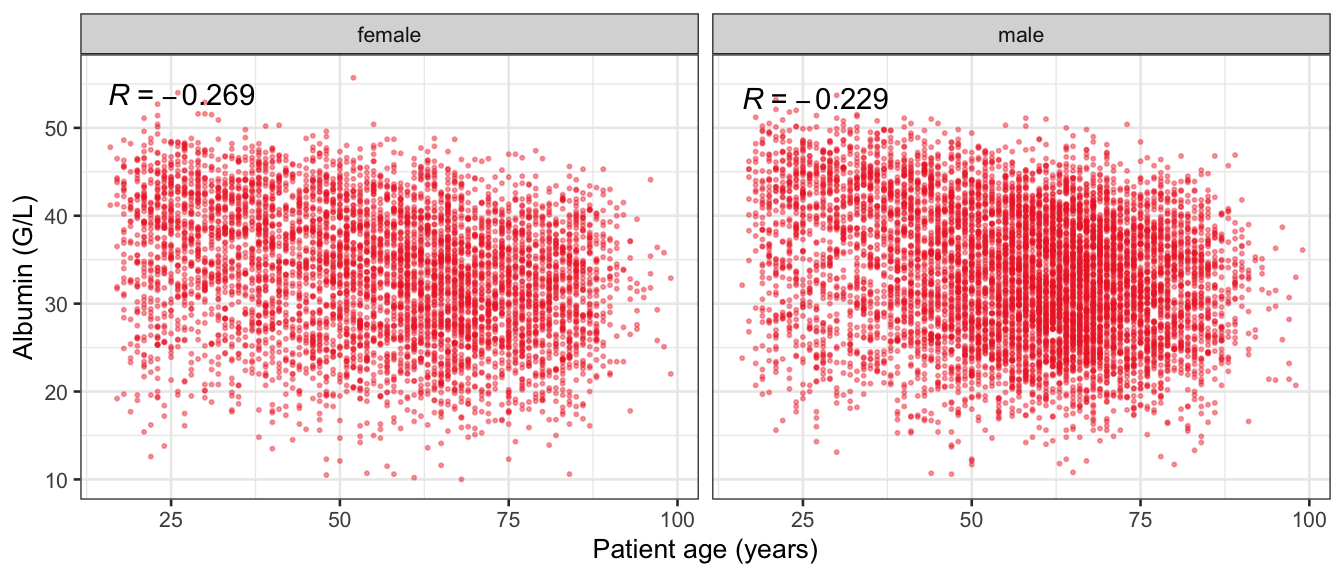
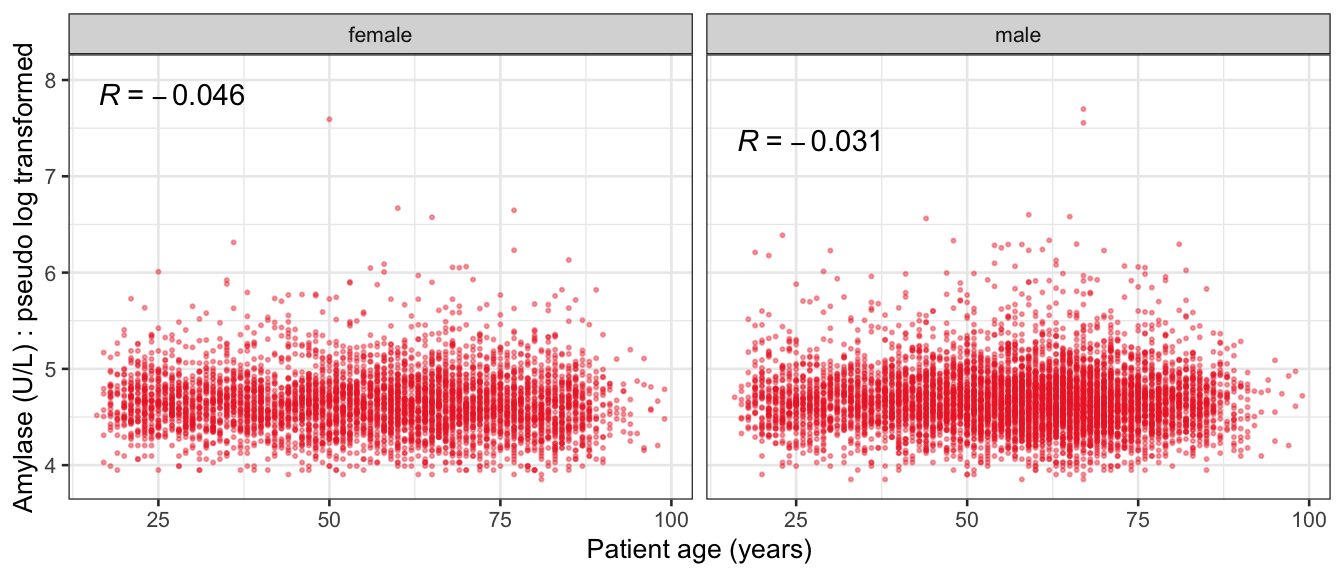
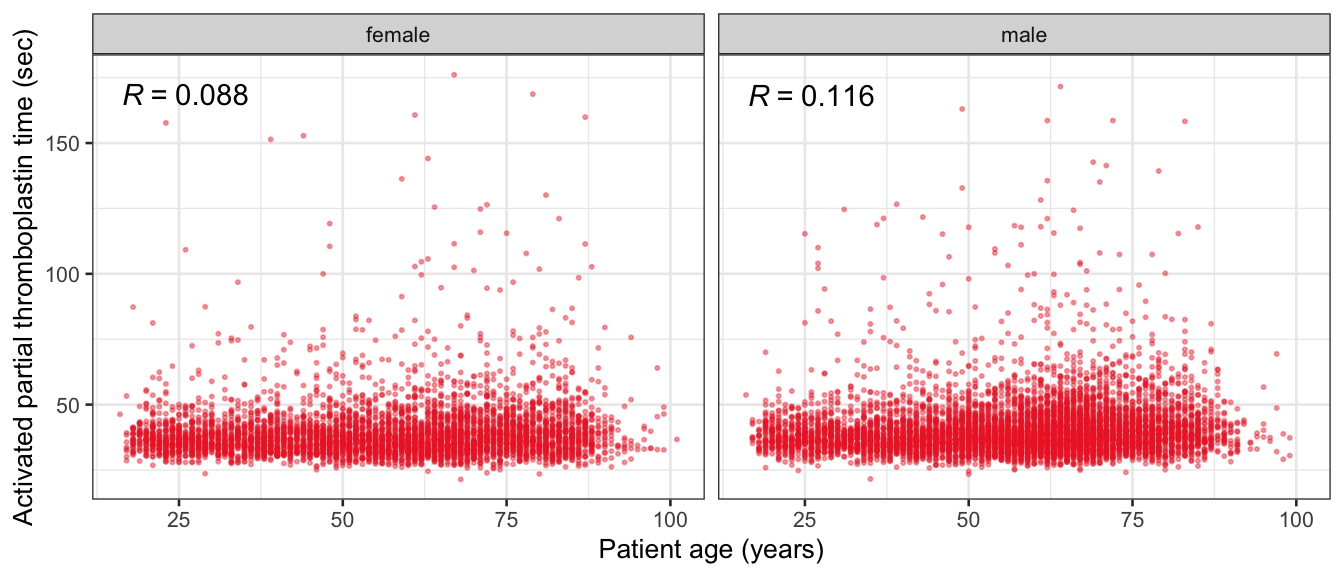
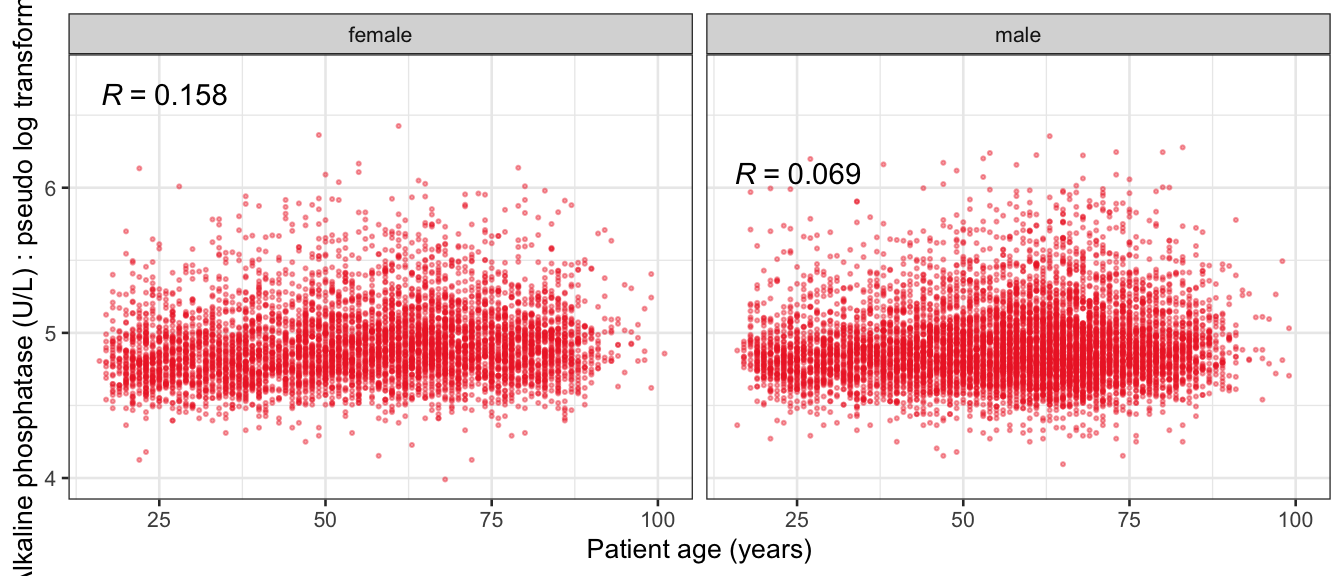
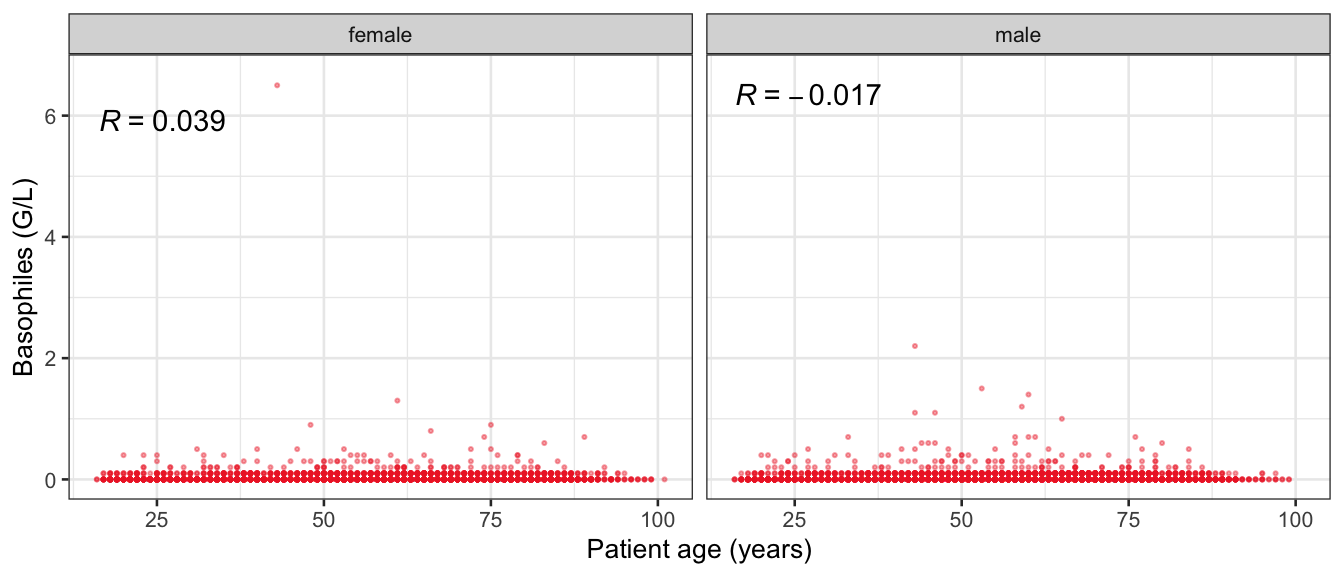
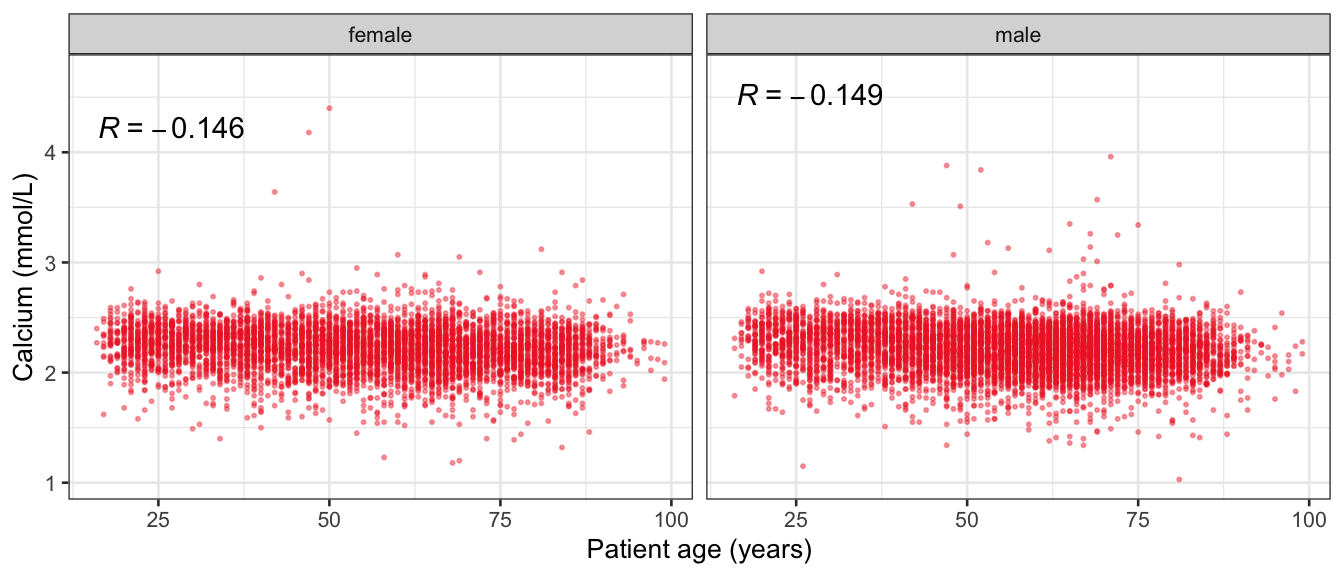
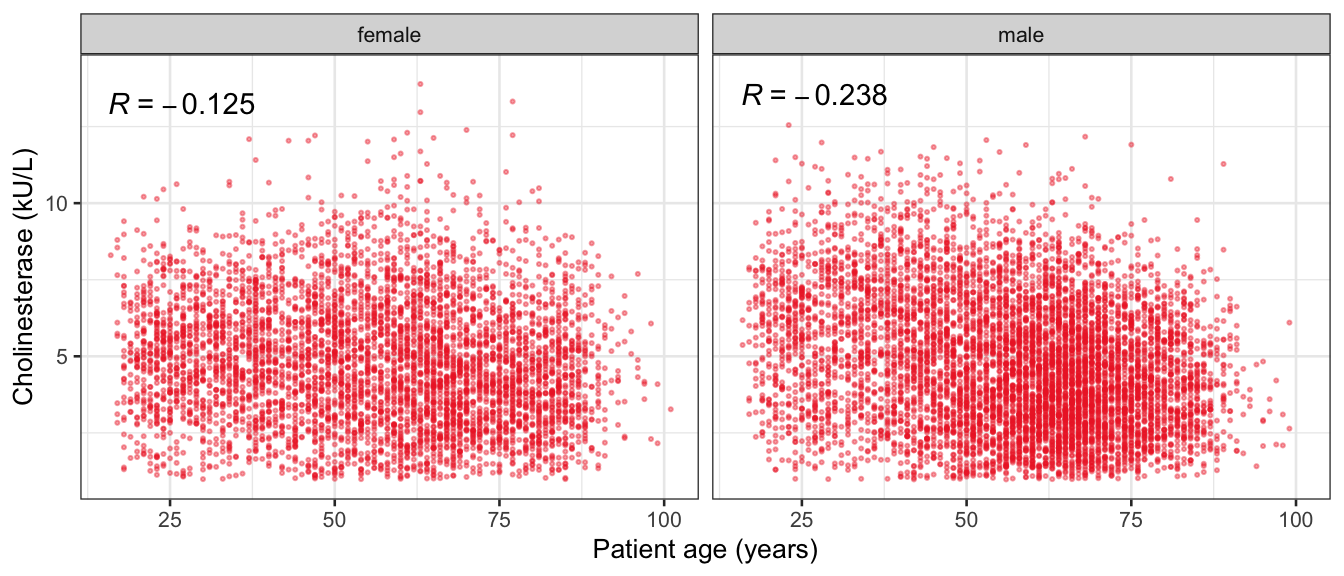
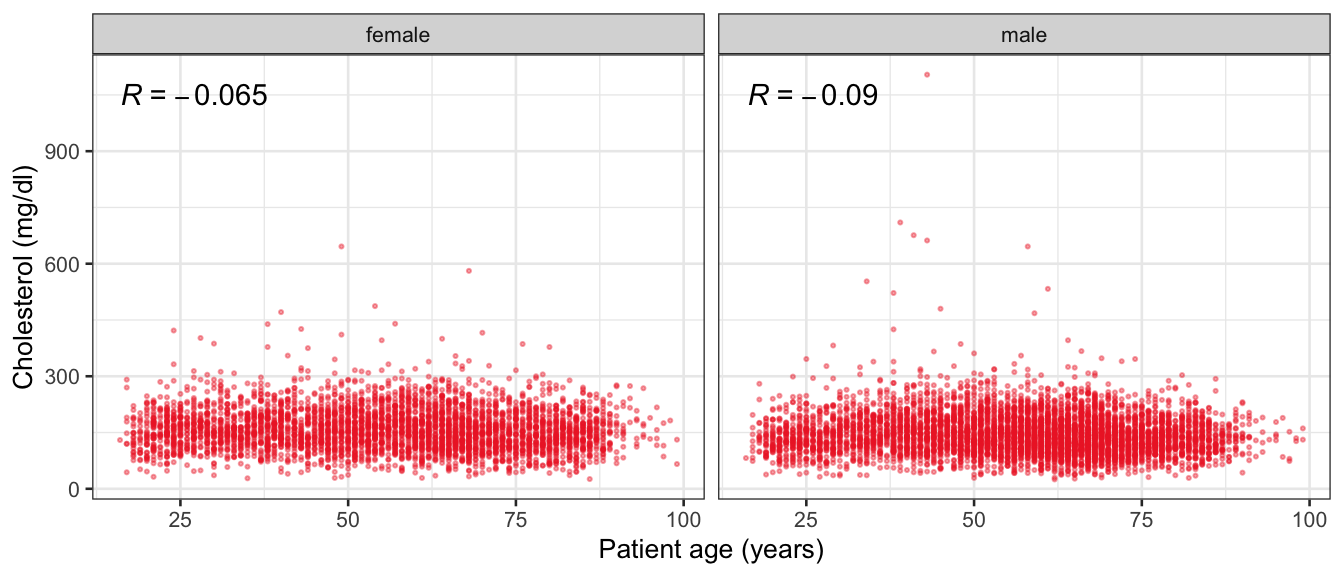
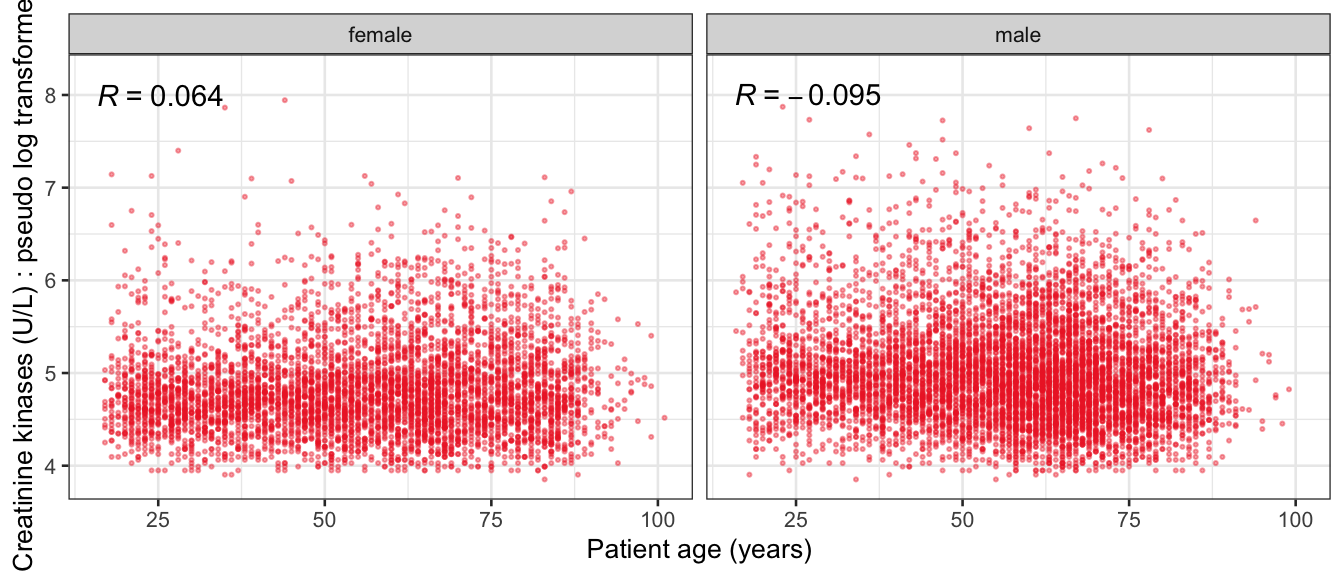
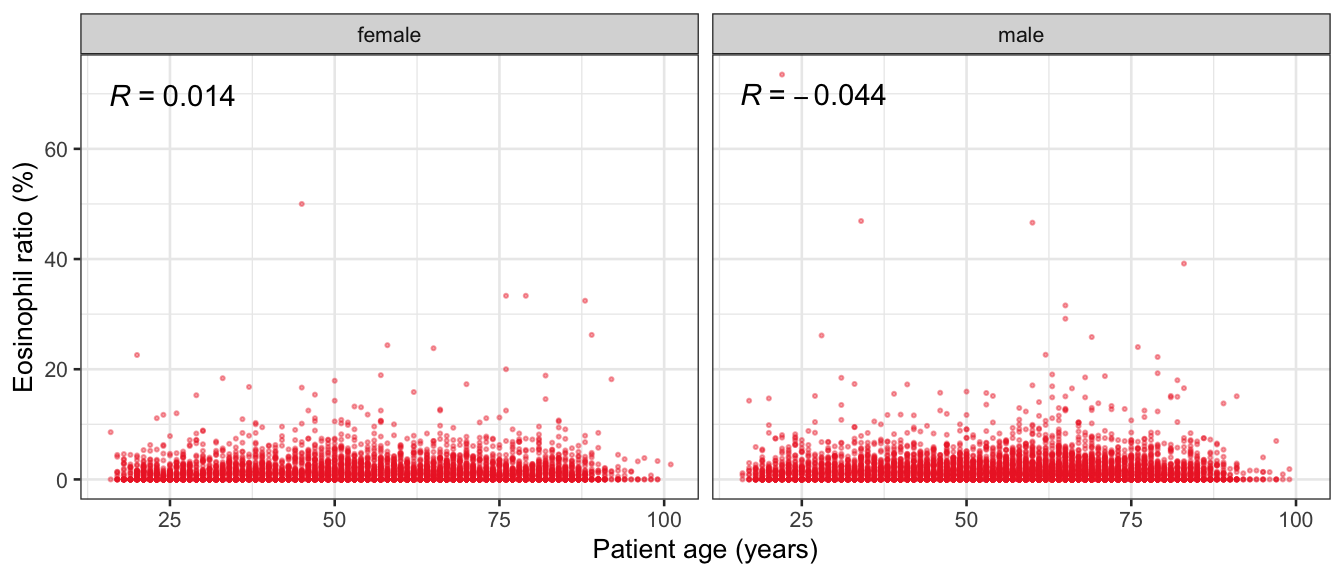
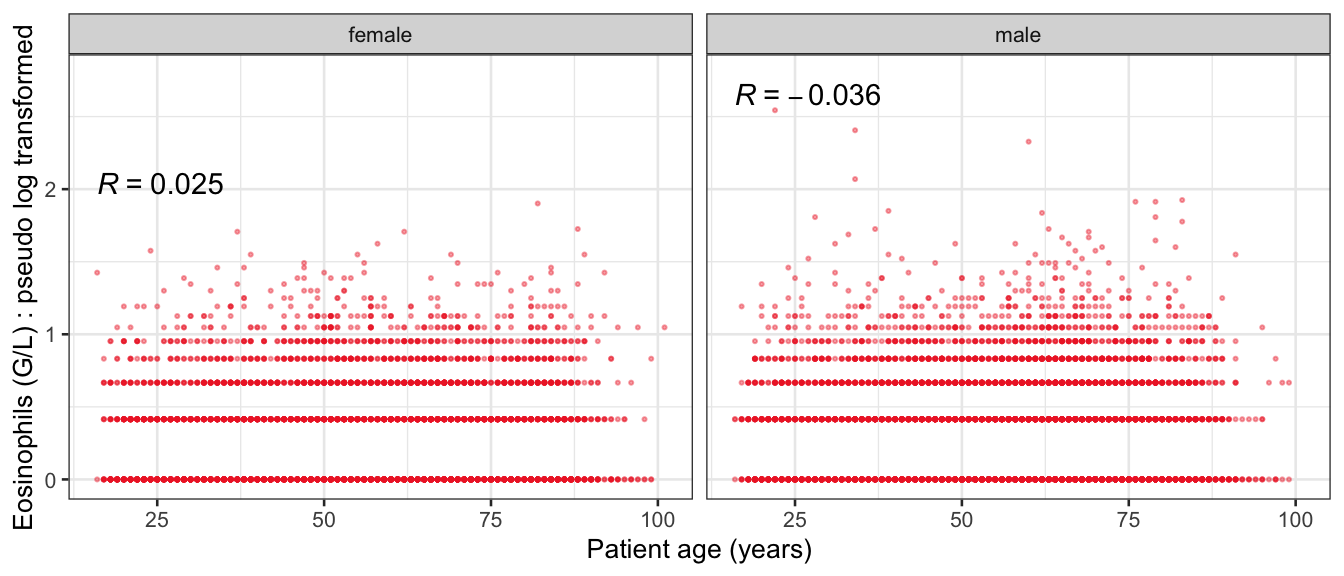
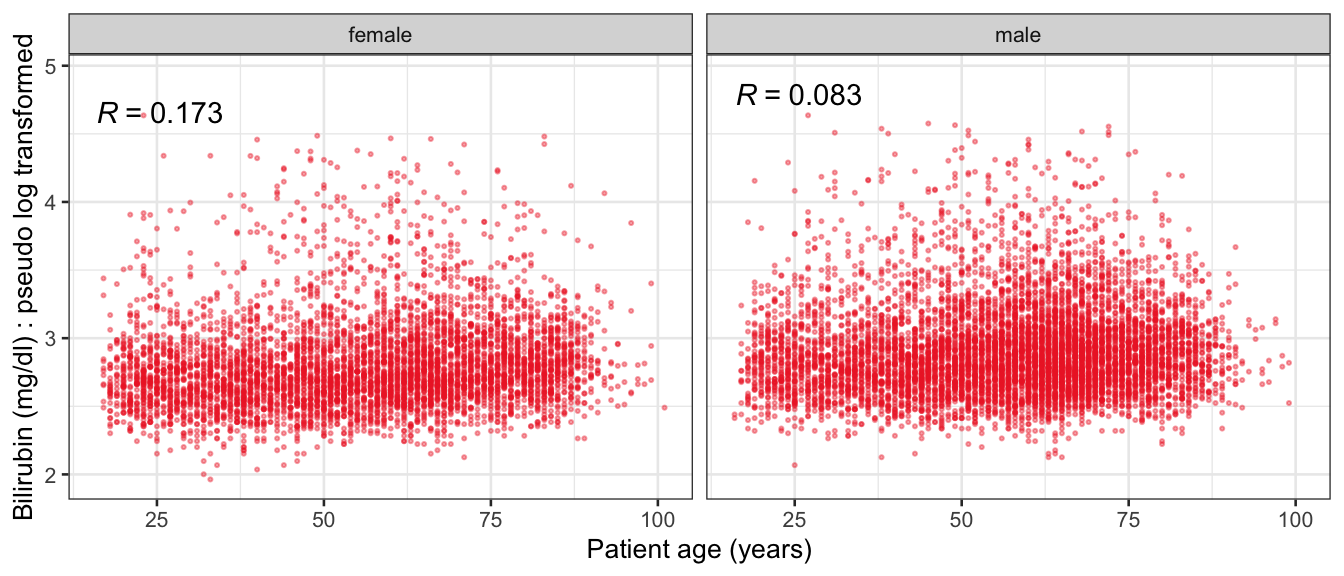
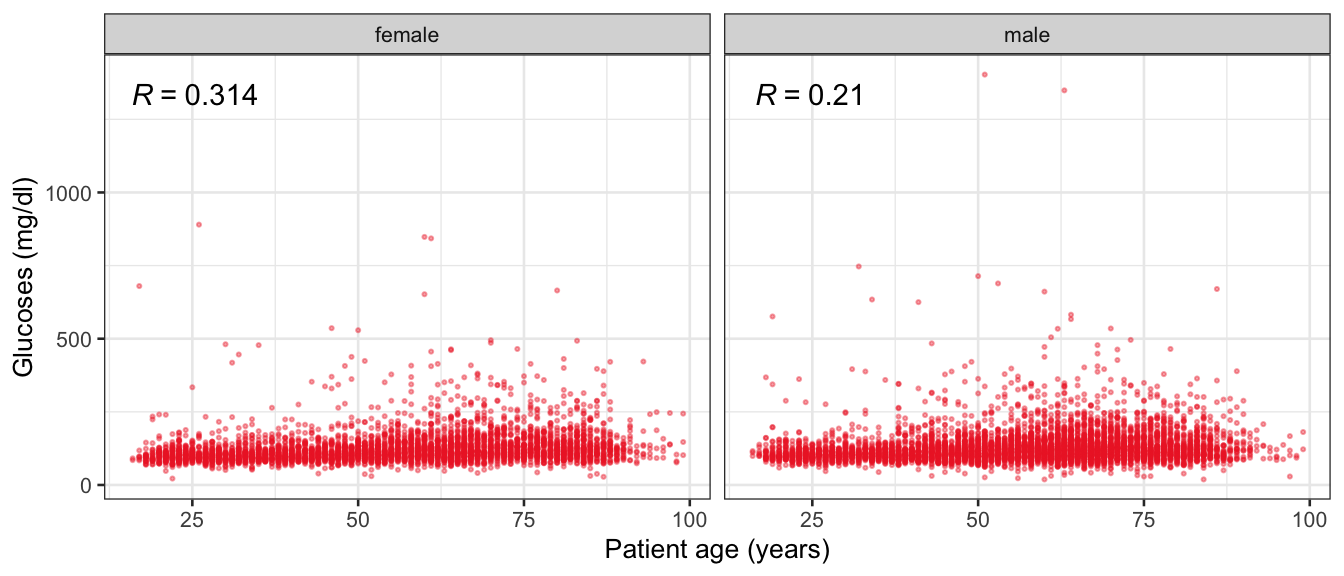
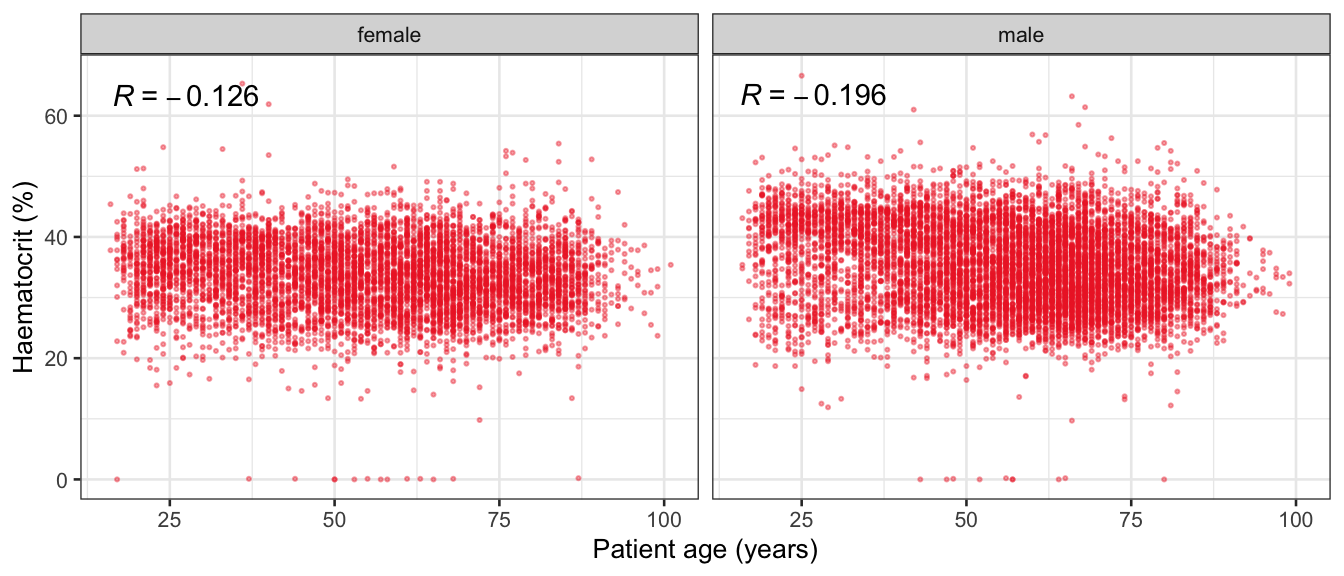
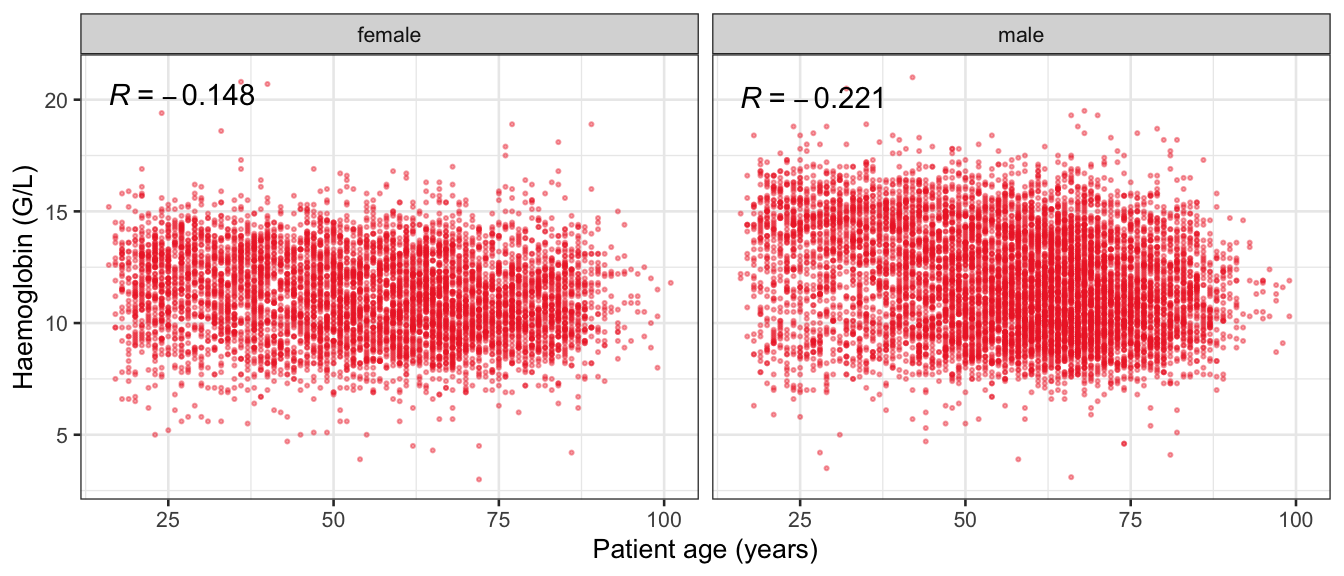
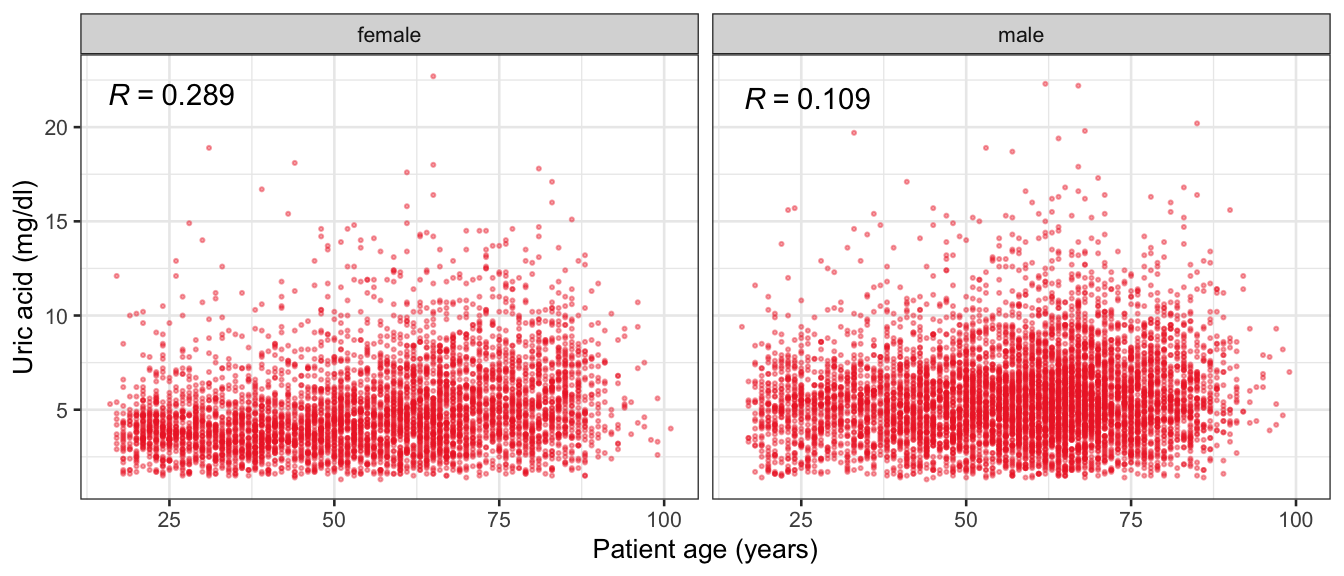
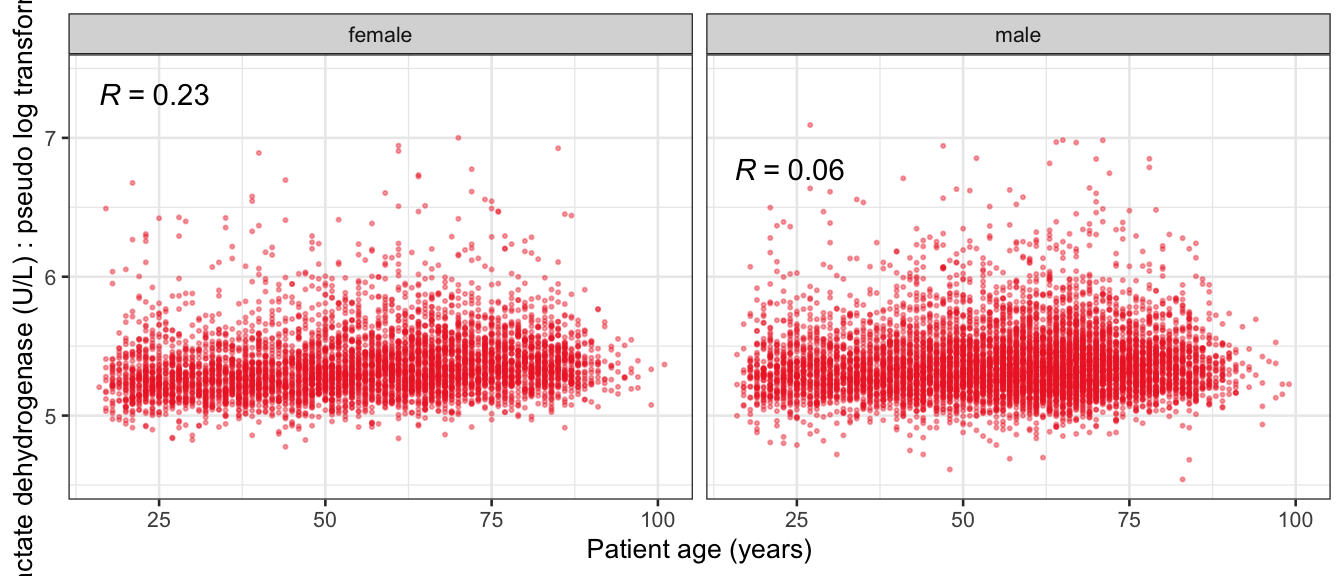
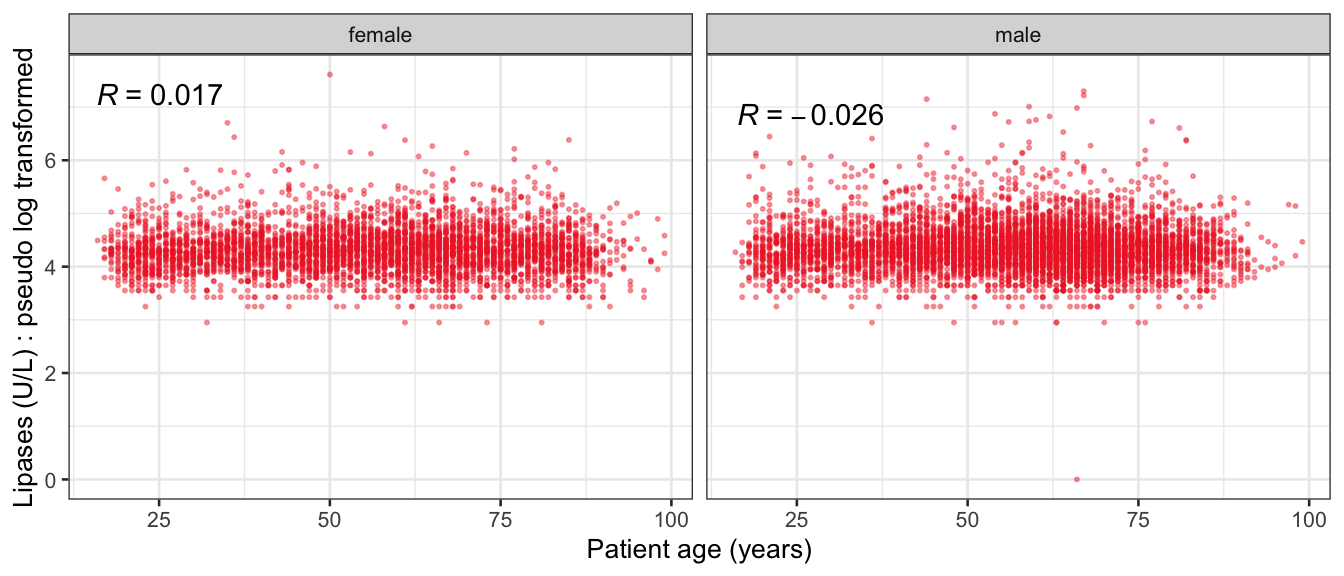
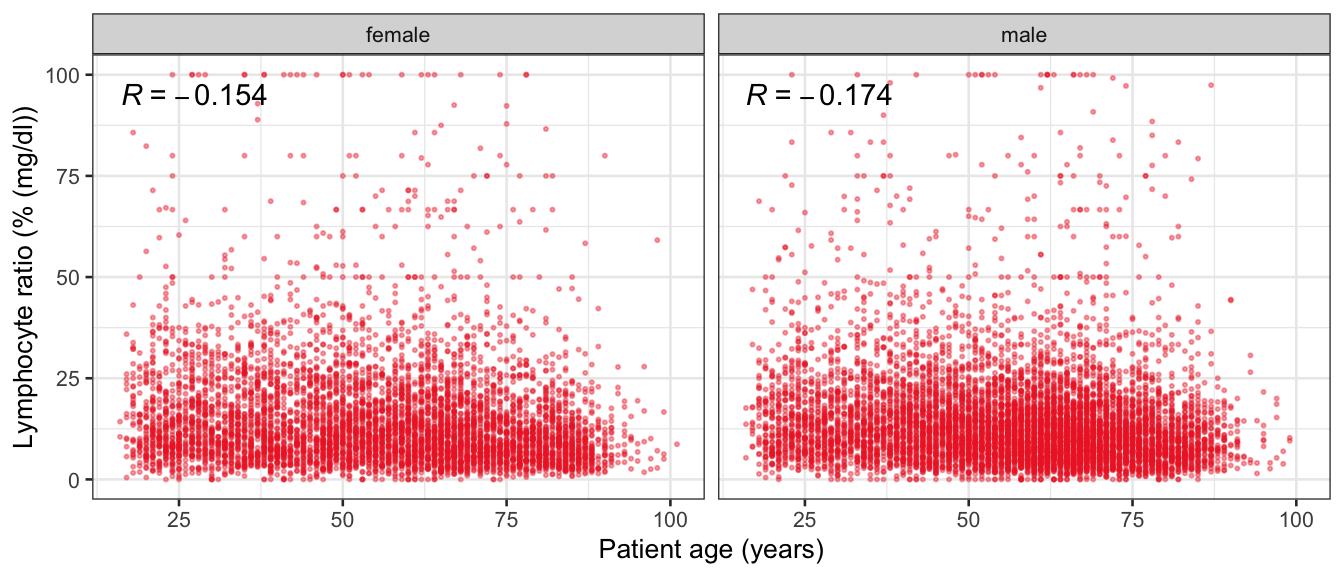
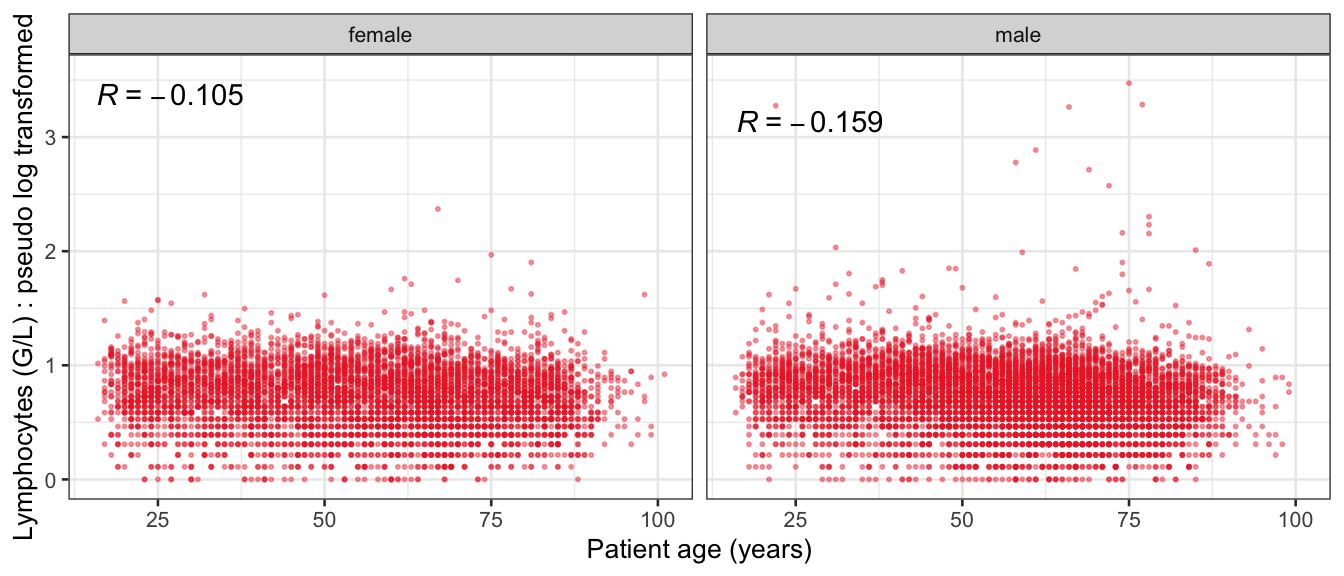
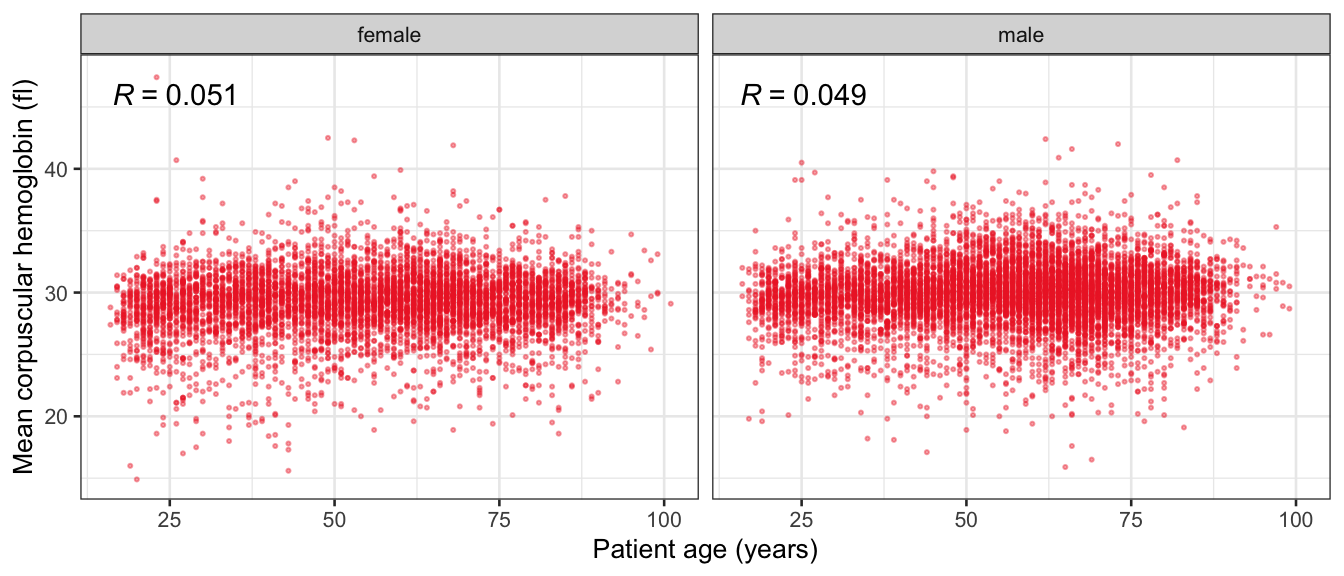
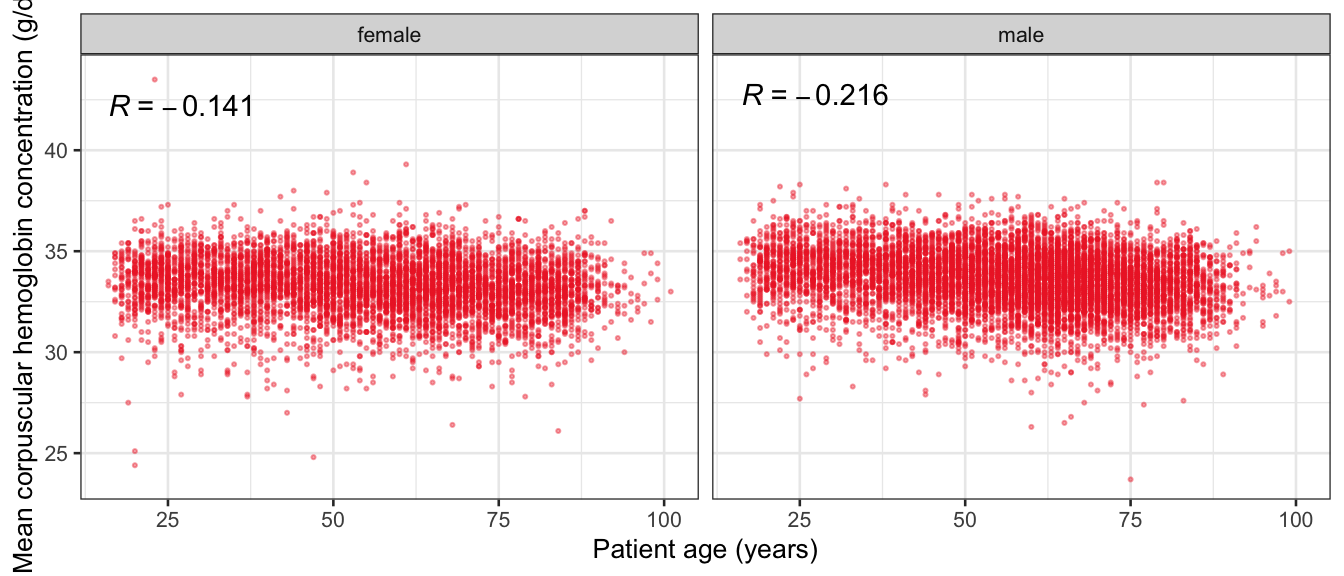
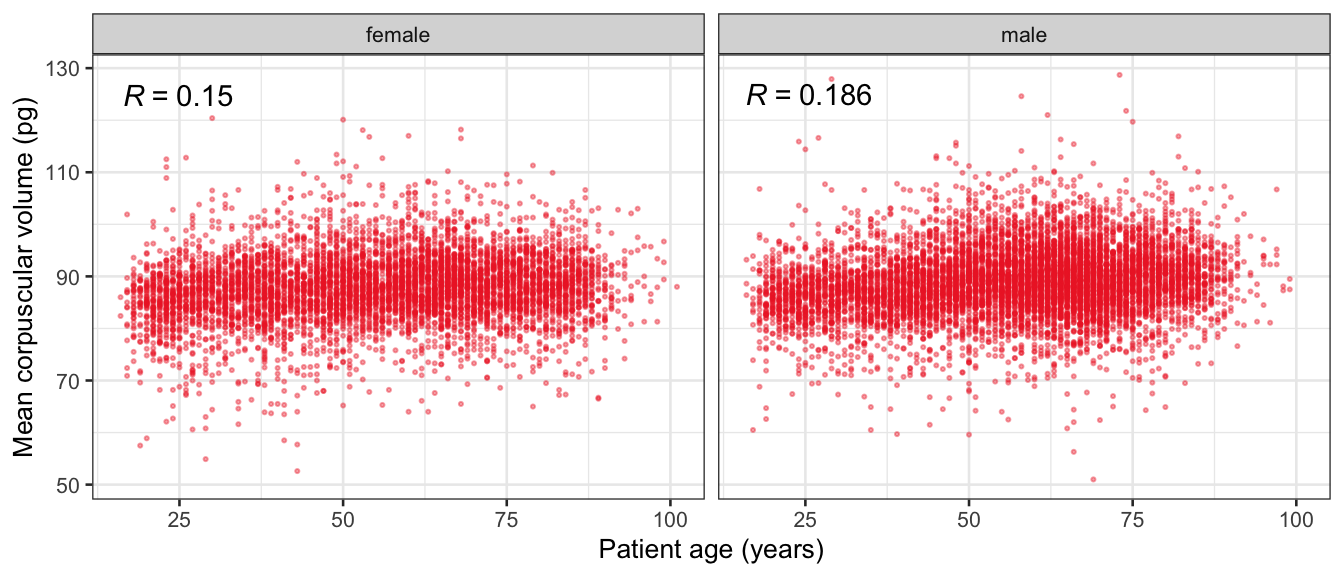
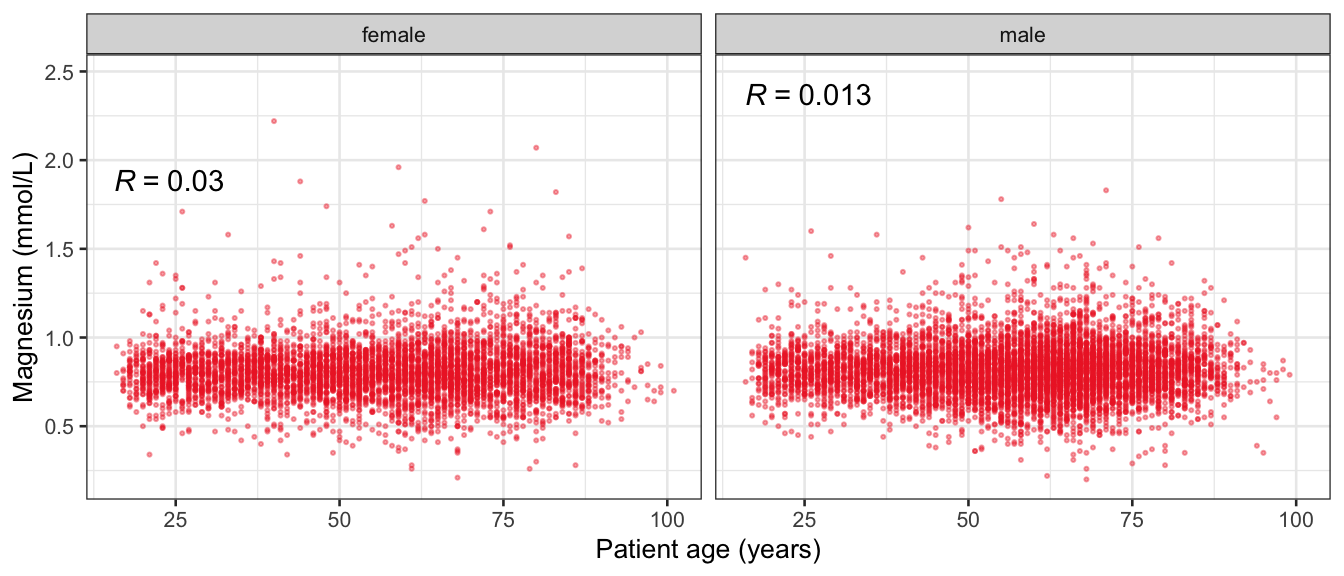
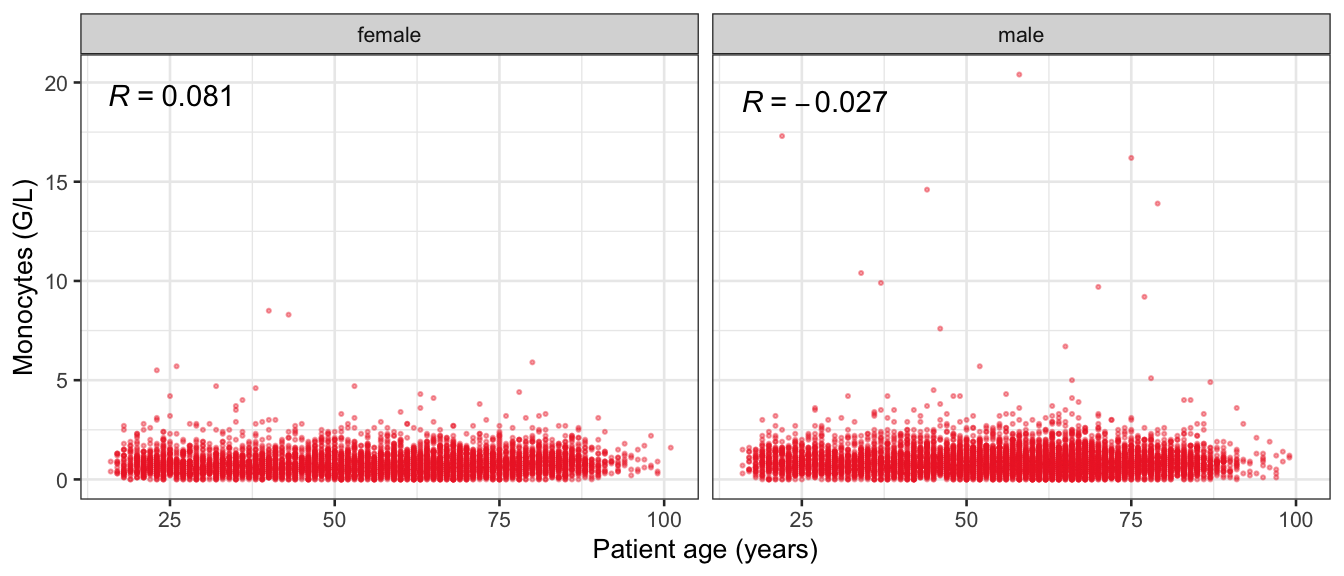
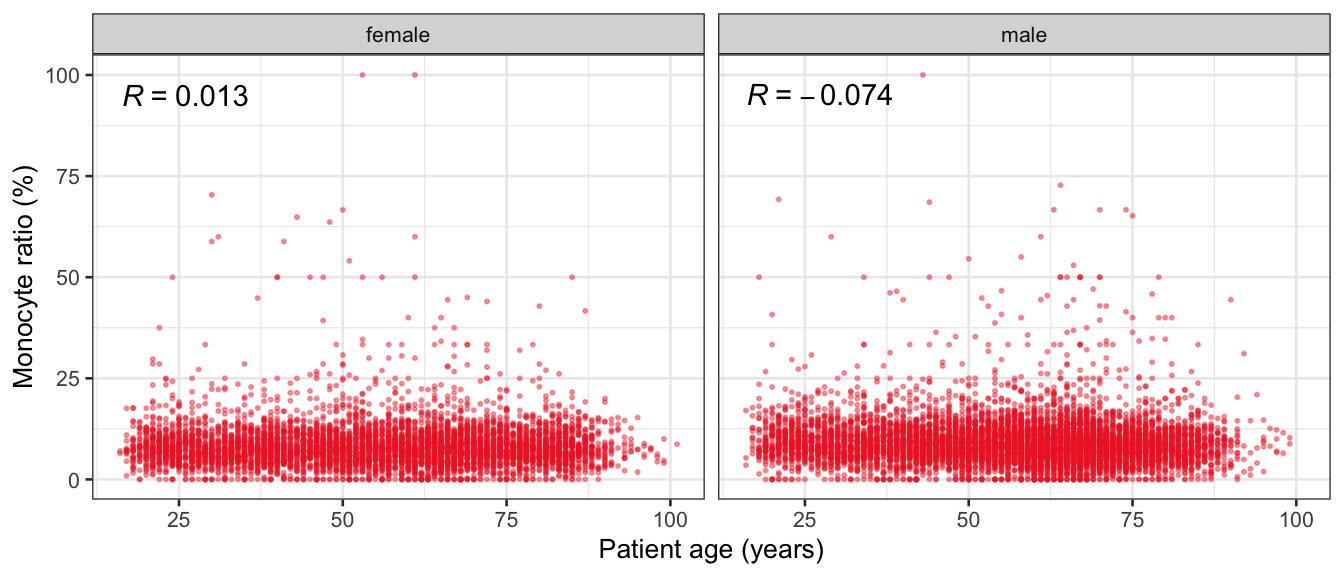
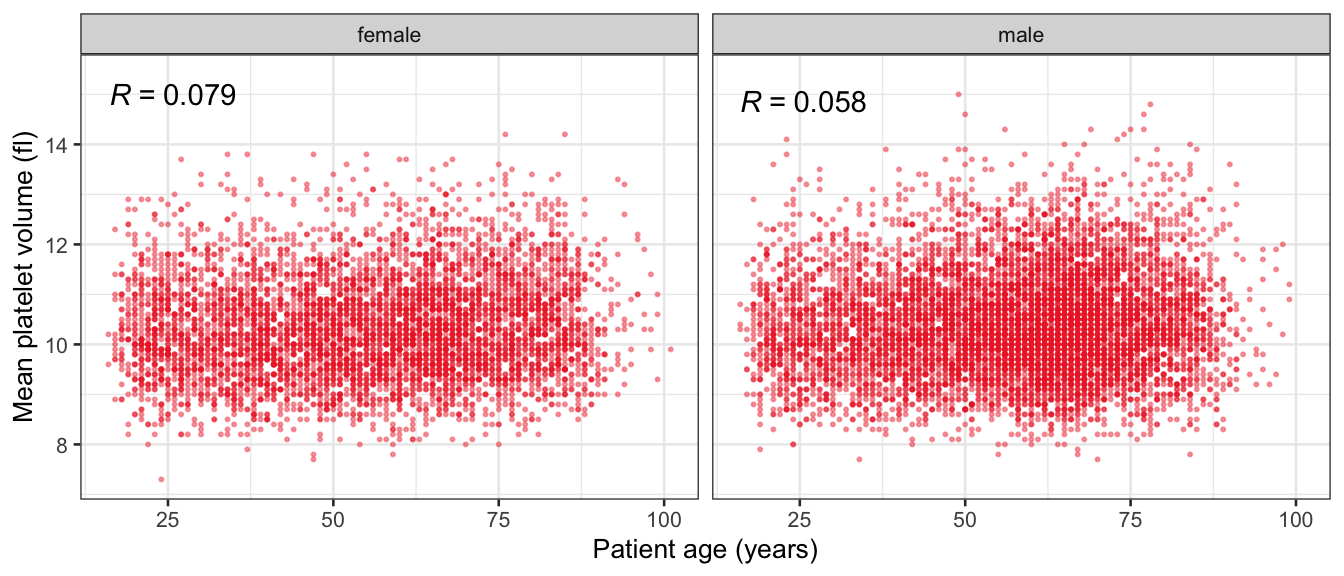
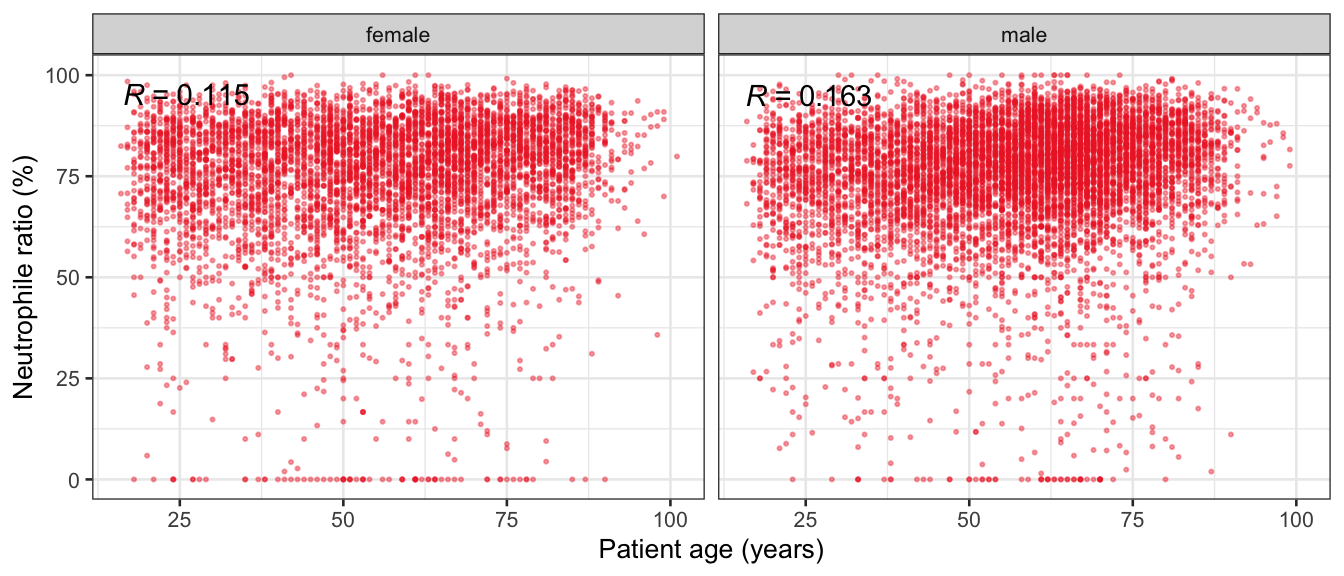
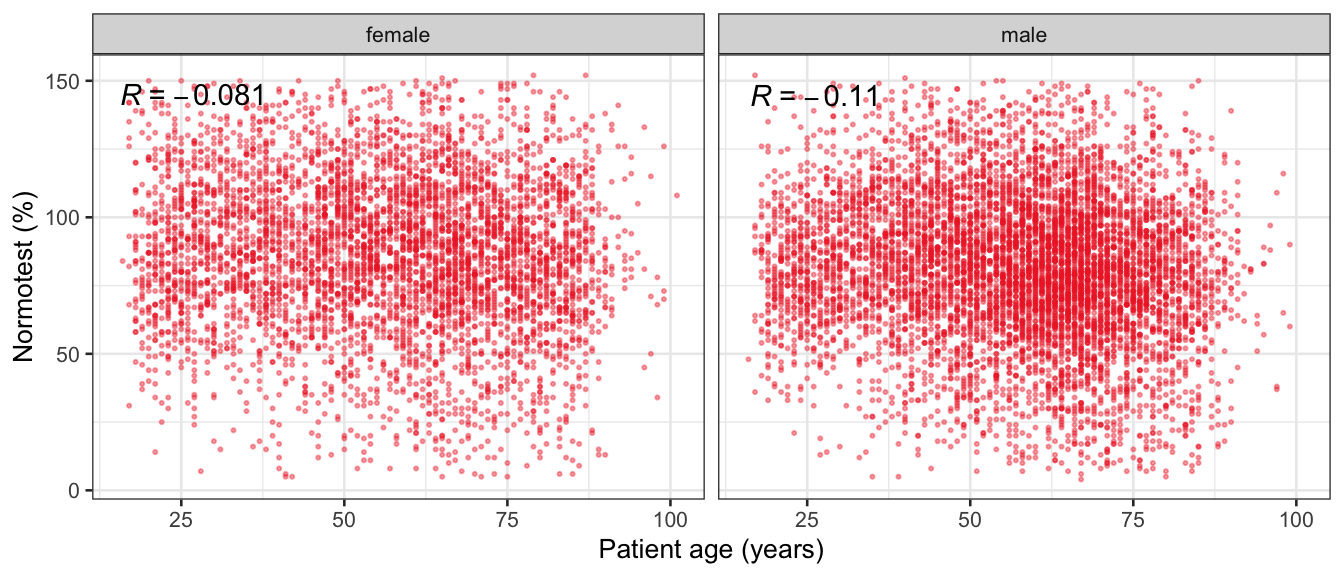
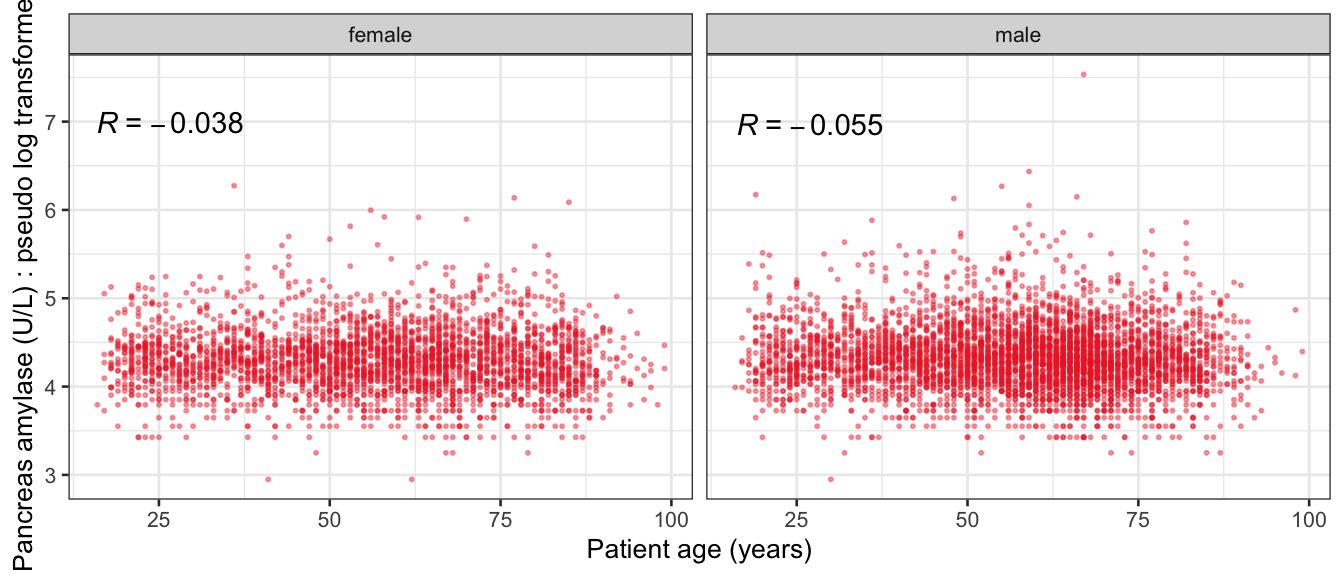
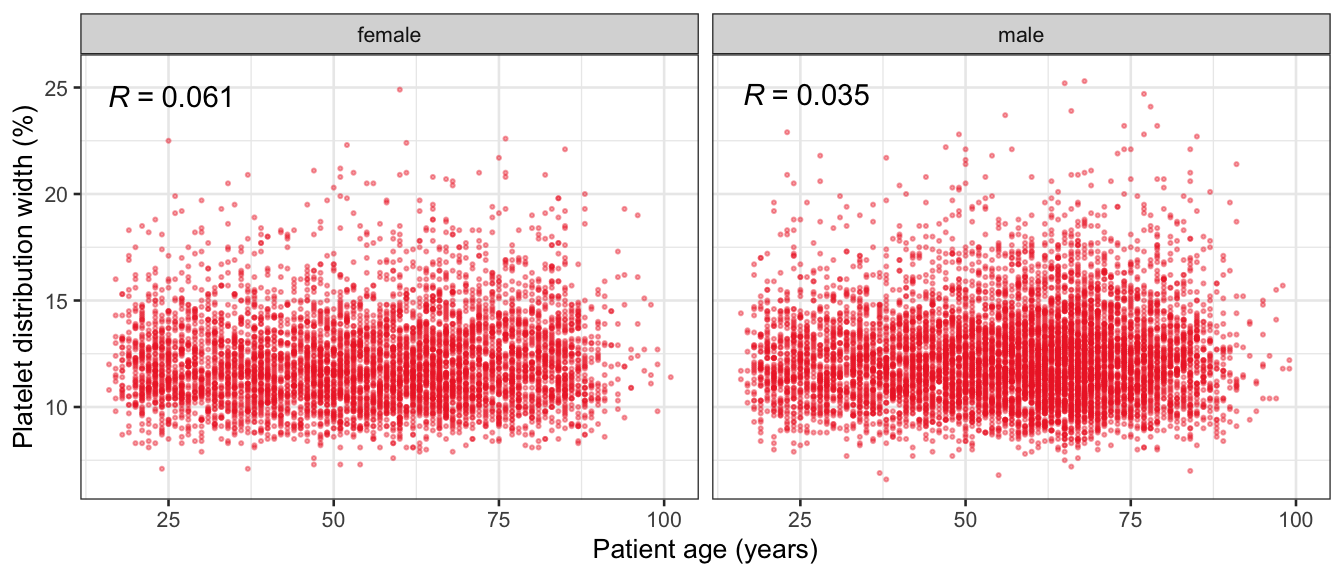
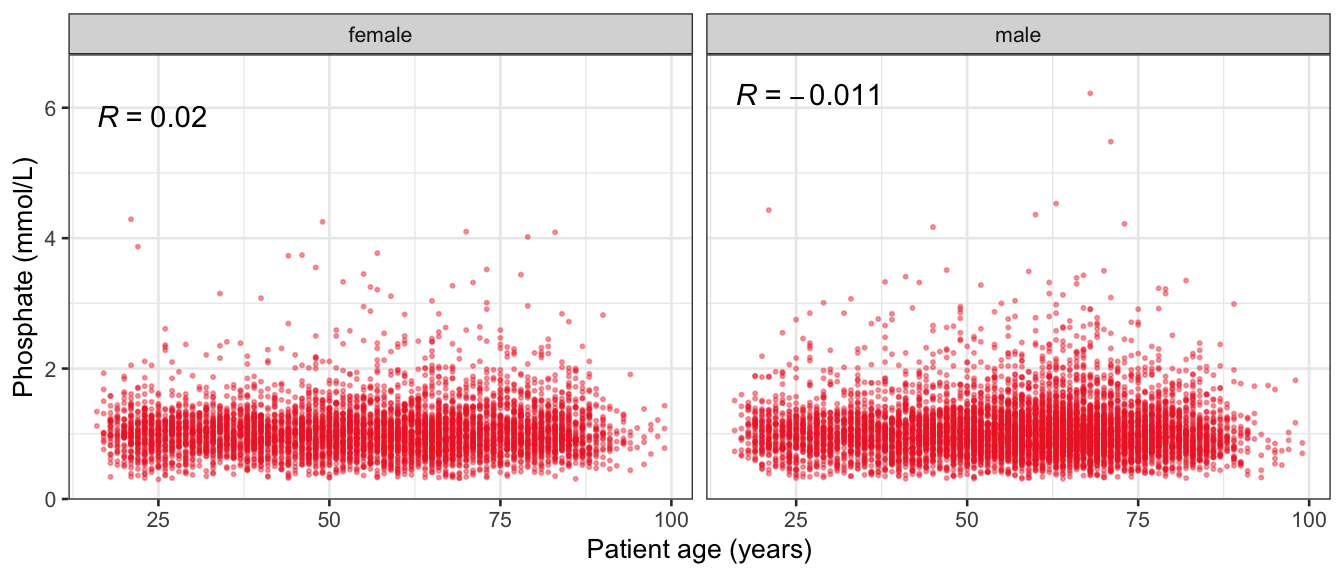
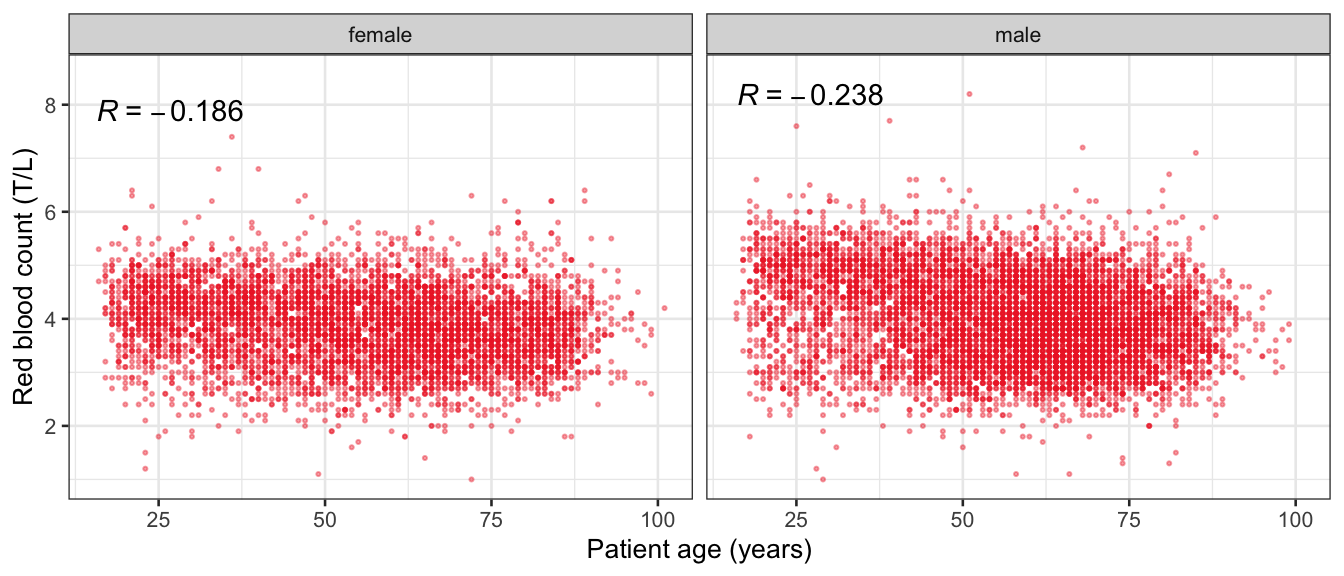
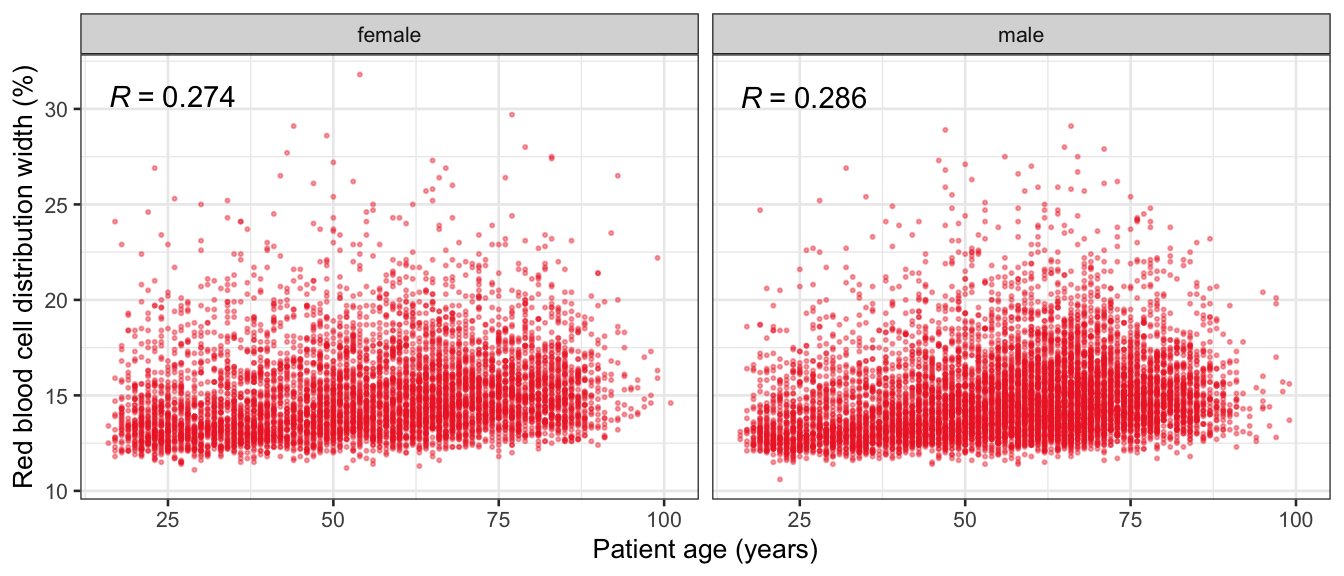
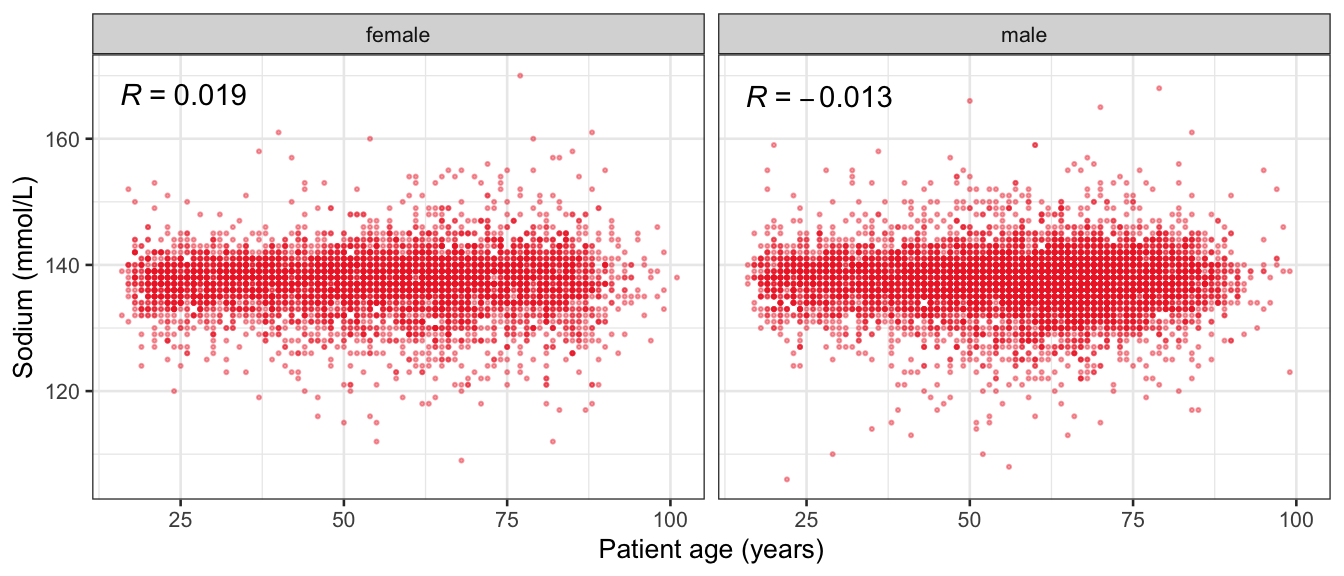
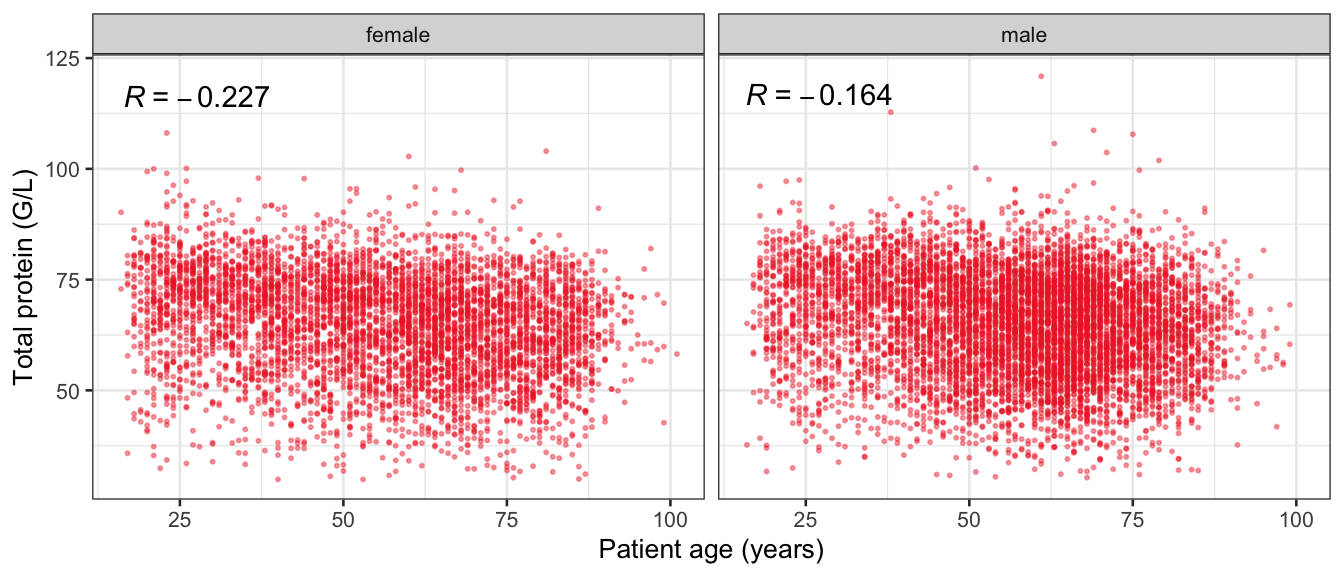
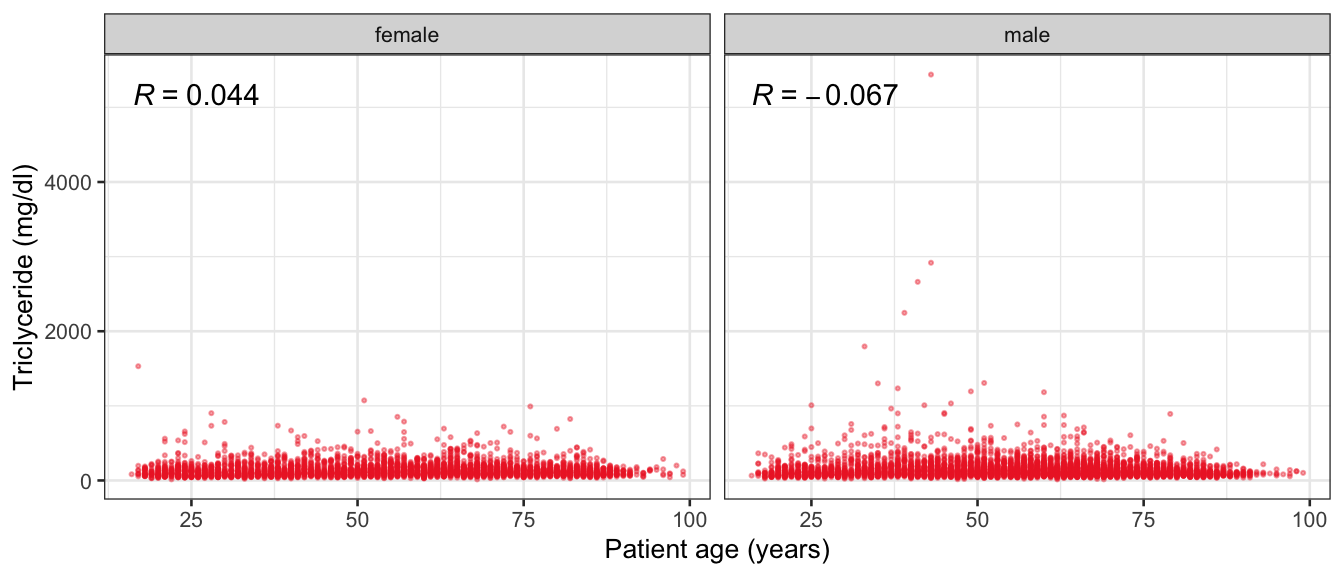
# Multivariate analyses {#sec-multivar-appendix} `data/IDA/ADLB_02.rds` . ```{r, echo = FALSE, message = FALSE, warning = FALSE } library (tidyr)library (dplyr)library (ggplot2)library (purrr)library (tidyselect)library (corrr) ## tidy correlation library (patchwork)library (ggpubr)library (ggrepel)library (here)library (dendextend)library (gt)library (gtExtras)## for network plot options (ggrepel.max.overlaps = 100 )source (here ("R" , "fun_compare_dist_plot.R" ))source (here ("R" , "fun_compare_mult_dists.R" ))source (here ("R" , "fun_ida_trans.R" ))## Load the datasets - make sure to load the correct ADLB2 ## TODO : have a make workflow to ensure dependencies run in correct order <- readRDS (here:: here ("data" ,"IDA" , "ADSL_01.rds" ))<- readRDS (here:: here ("data" ,"IDA" , "ADLB_02.rds" ))``` ## V1: Association with structural variables {#sec-V1-appendix} ```{r mvi01a, message=FALSE, warning=FALSE, echo=FALSE, fig.height=3} #| layout-ncol: 3 ## Join with ADSL for structural variables <- |> select (USUBJID, AGEGR01C, AGE, SEXC, SEX) <- ADLB |> left_join (ADSL01, by = "USUBJID" )<- dat |> filter ( REM_PRED_FL02 == "Y" ) |> group_by (PARAMCD) |> group_map (~ plot_assoc_by (.x), .keep = TRUE )for (plts in remain_predictors) {print (plts)``` ## VE3: Redundancy {#sec-VE3-appendix} ### Linear model ```{r vif_app, message=FALSE, warning=FALSE, echo=FALSE, fig.height=3} ## Join with ADSL for structural variables <- |> select (USUBJID, AGEGR01C, AGE, SEXC, SEX) <- ADLB |> left_join (ADSL01, by = "USUBJID" )<- dat |> filter (KEY_PRED_FL02 == "Y" | MED_PRED_FL02 == "Y" | REM_PRED_FL02 == "Y" ) |> select (USUBJID, PARAMCD, AVAL, SEX) |> pivot_wider (names_from = PARAMCD, values_from = AVAL, values_fill = NA ) <- dat_wide |> select (- USUBJID)<- as.formula (paste (c ("~" ,paste (names (dat_vif), collapse= "+" )), collapse= "" ))#formula <- Hmisc:: redun (formula, data = dat_vif, nk= 0 , pr= FALSE )<- 1 / (1 - red$ rsq1)#cat("\nAvailable samvple size:\n", red$n, " (", #round(100*red$n/nrow(dat),2), "%)\n") <- as_tibble (vif, rownames = "PARAMCD" ) |> rename (vif = value)<- as_tibble (red$ rsq1, rownames = "PARAMCD" ) |> rename (r2 = value)<- vif_df |> left_join (red_df, by = "PARAMCD" )``` `r red$n` (`r round(100*red$n/nrow(dat),2)` %).```{r vif_app02, message=FALSE, warning=FALSE, echo=FALSE, fig.height=3} #vif_df |> gt::gt(caption = "Variance inflation factors") #red_df |> gt::gt(caption = "Multiple R-squared") |> gt () |> :: cols_label (PARAMCD = "Parameter code" ,r2 = "Multiple R-squared" ,vif = "Variance inflation factor" |> fmt_number (columns = c (r2),decimals = 2 |> fmt_number (columns = c (vif),decimals = 1 |> gt_theme_538 ()```