Code
pseudo_log <-
function(x, sigma = 1, base = 10)
asinh(x / (2 * sigma)) / log(base)
inv_pseudo_log <-
function(x, sigma = 1, base = 10)
2 * sigma * sinh(x * log(base))This supplemental section illustrates how pseudo-log transformations can be used to transform skewed distributions towards normality.
The transformation \(f(x) = sinh^-1(x/2\sigma)/log(10)\) is a pseudo-logarithmic transformation mentioned by (Johnson 1949). It has the following advantages over ordinary logarithmic transformations:
Of course, this comes at the cost of deviation from the logarithmic transformation in terms of interpretability.
The parameter \(\sigma\) may be used to adapt the transformation to a specific range of an empirical distribution.
We define the pseudo-logarithmic transformation in R as:
pseudo_log <-
function(x, sigma = 1, base = 10)
asinh(x / (2 * sigma)) / log(base)
inv_pseudo_log <-
function(x, sigma = 1, base = 10)
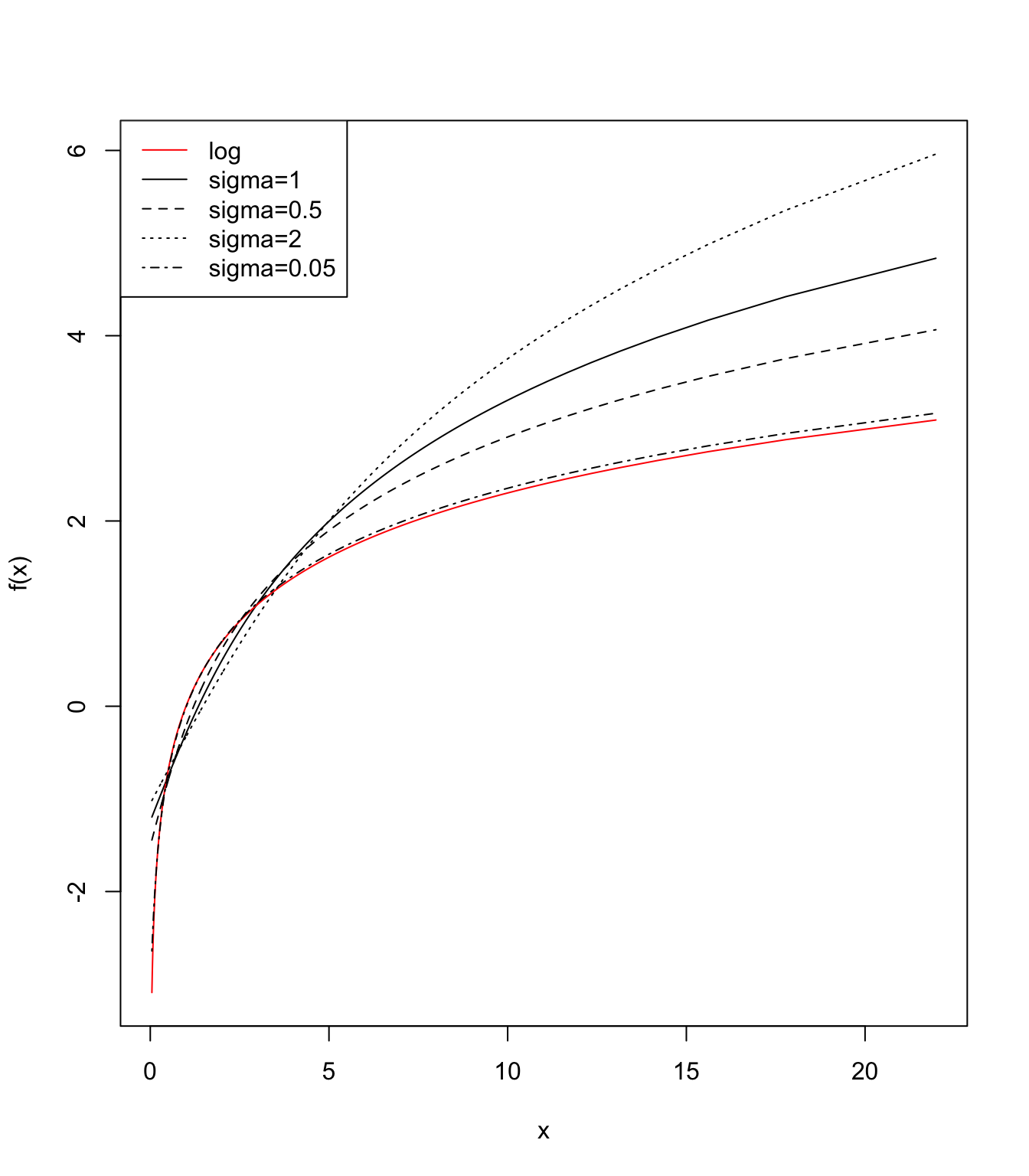
2 * sigma * sinh(x * log(base))Next, we investigate how the parameter \(\sigma\) impacts the result of the transformation. We assume that \(x\) follows a log-normal distribution, and we will show results of \(f(x)\) with different choices of \(\sigma\). We center and scale \(f(x)\) before plotting.
p <- seq(0.001, 0.999, 0.001)
x <- exp(qnorm(p, mean = 0, sd = 1))
hist(x)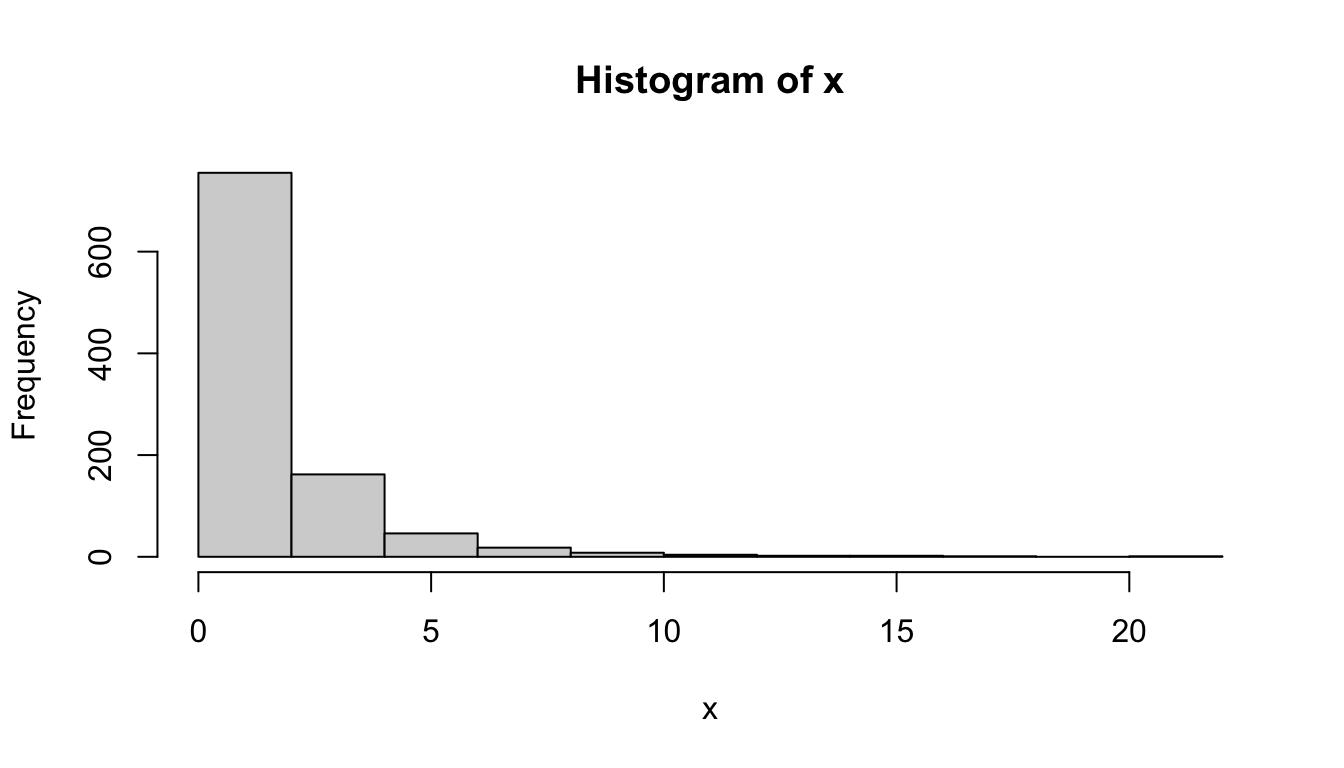
y <-
cbind(
log(x),
scale(pseudo_log(x, 1)),
scale(pseudo_log(x, 0.5)),
scale(pseudo_log(x, 2)),
scale(pseudo_log(x, 0.05))
)
plot(x,
y[, 2],
type = "l",
ylab = "f(x)",
ylim = range(y))
lines(x, y[, 1], col = "red")
lines(x, y[, 3], type = "l", lty = 2)
lines(x, y[, 4], type = "l", lty = 3)
lines(x, y[, 5], type = "l", lty = 4)
legend(
"topleft",
lty = c(1, 1, 2, 3, 4),
col = c("red", "black", "black", "black", "black"),
legend = c("log", "sigma=1", "sigma=0.5", "sigma=2", "sigma=0.05")
)
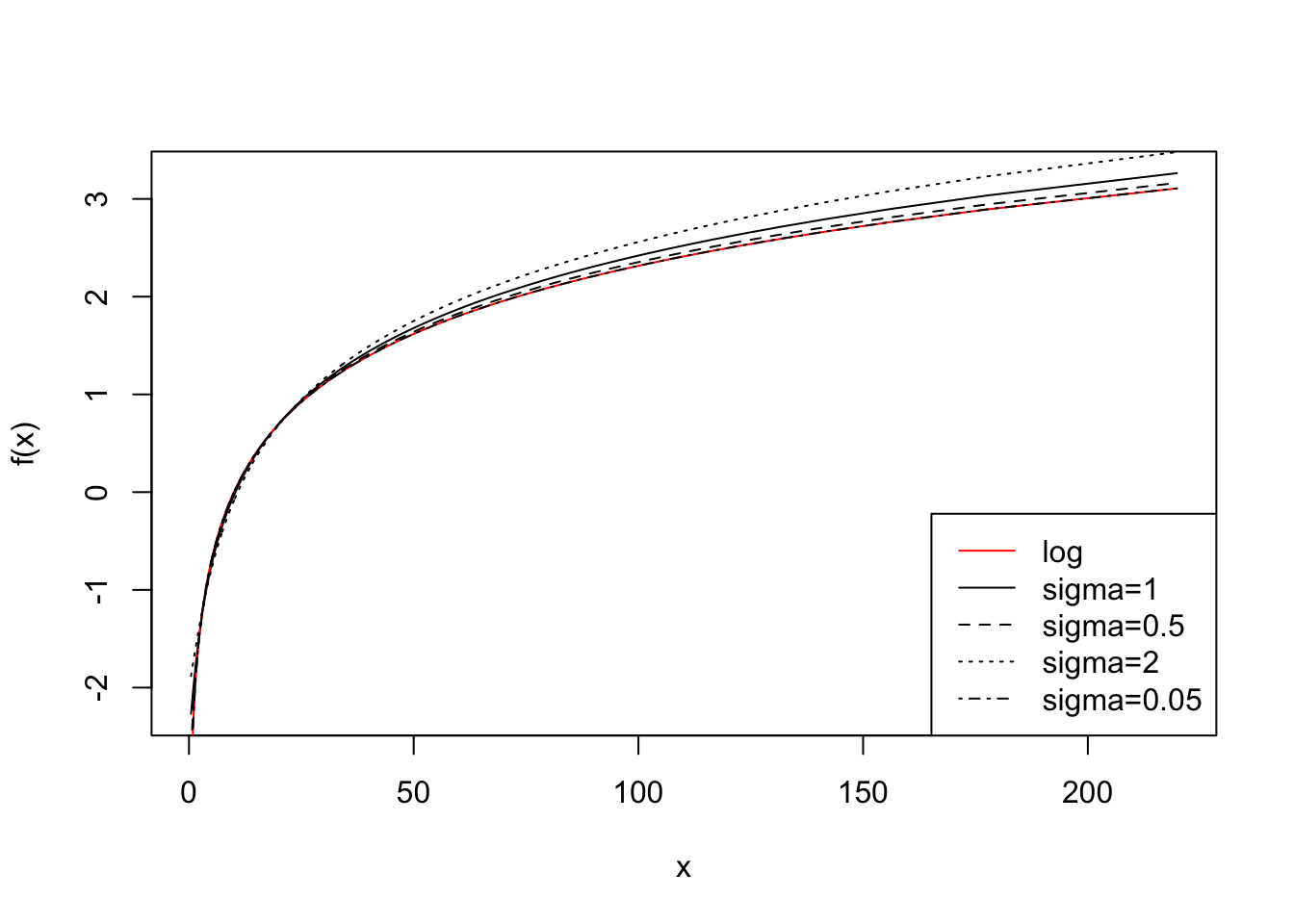
In the next code chunk, we multiply by 10 and repeat the exercise. We learn that the transformations become more similar and the choice of \(\sigma\) less relevant.
x <- x * 10
y <-
cbind(
scale(log(x)),
scale(pseudo_log(x, 1)),
scale(pseudo_log(x, 0.5)),
scale(pseudo_log(x, 2)),
scale(pseudo_log(x, 0.05))
)
plot(x, y[, 2], type = "l", ylab = "f(x)")
lines(x, y[, 1], col = "red")
lines(x, y[, 3], type = "l", lty = 2)
lines(x, y[, 4], type = "l", lty = 3)
lines(x, y[, 5], type = "l", lty = 4)
legend(
"bottomright",
lty = c(1, 1, 2, 3, 4),
col = c("red", "black", "black", "black", "black"),
legend = c("log", "sigma=1", "sigma=0.5", "sigma=2", "sigma=0.05")
)
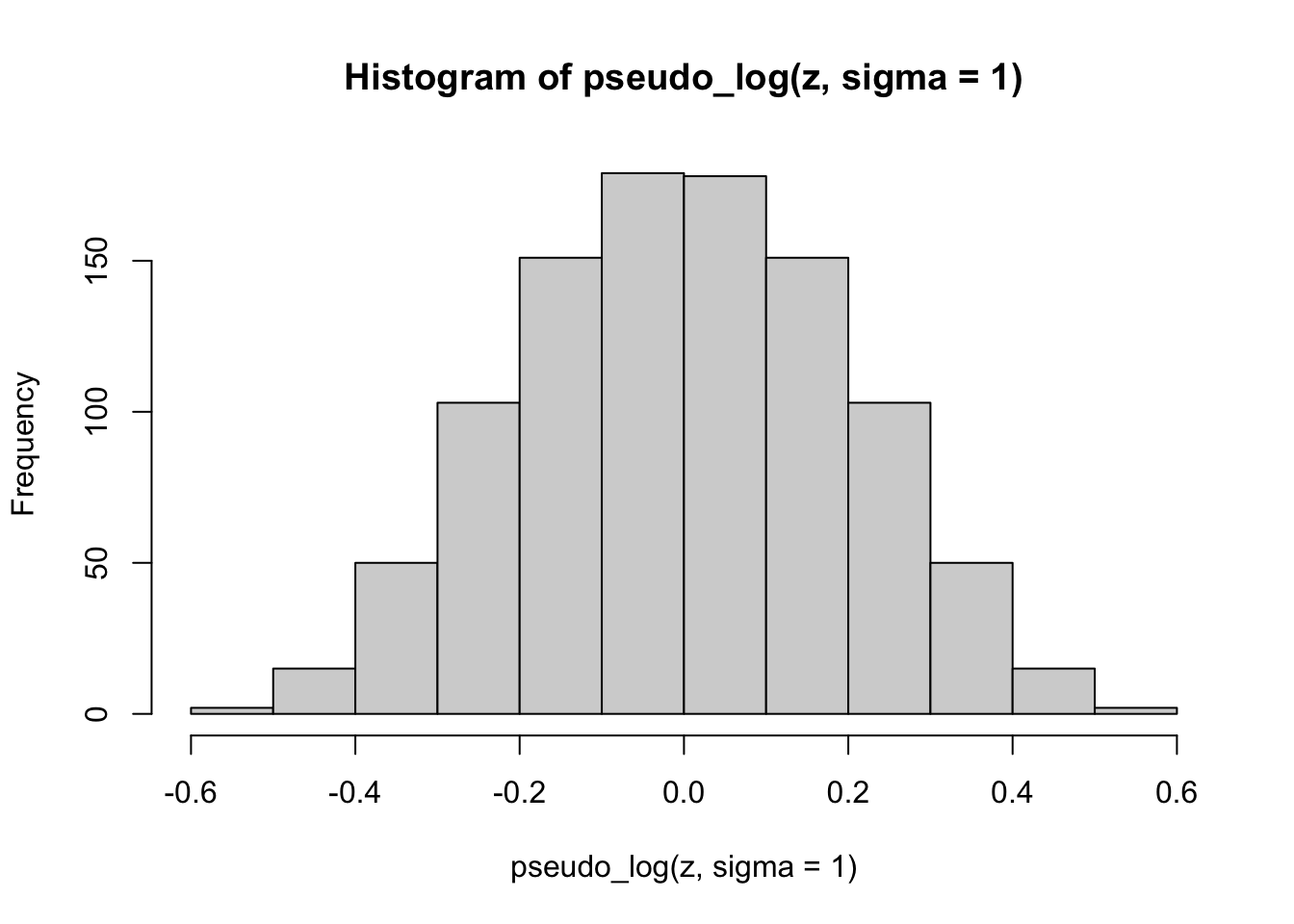
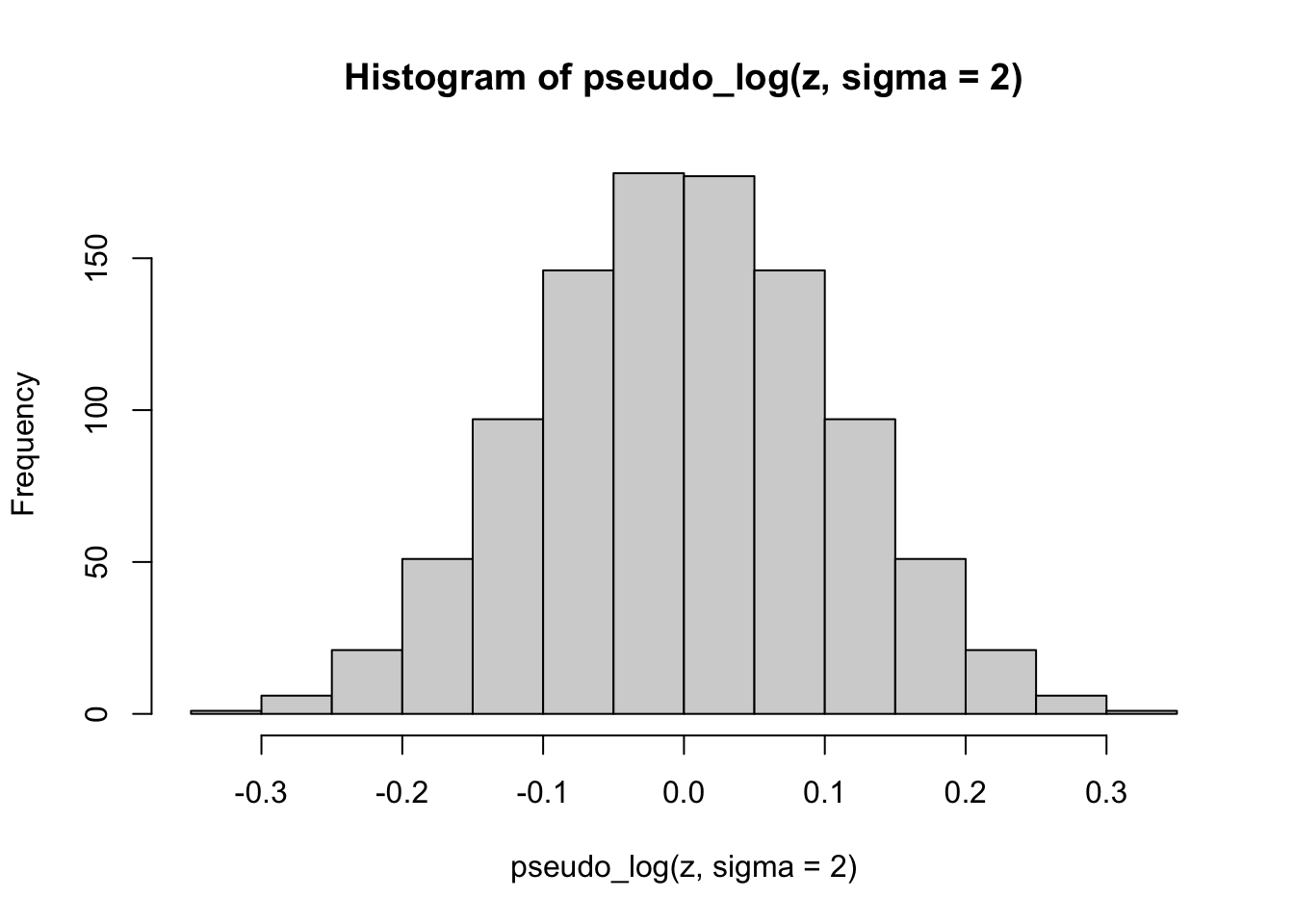
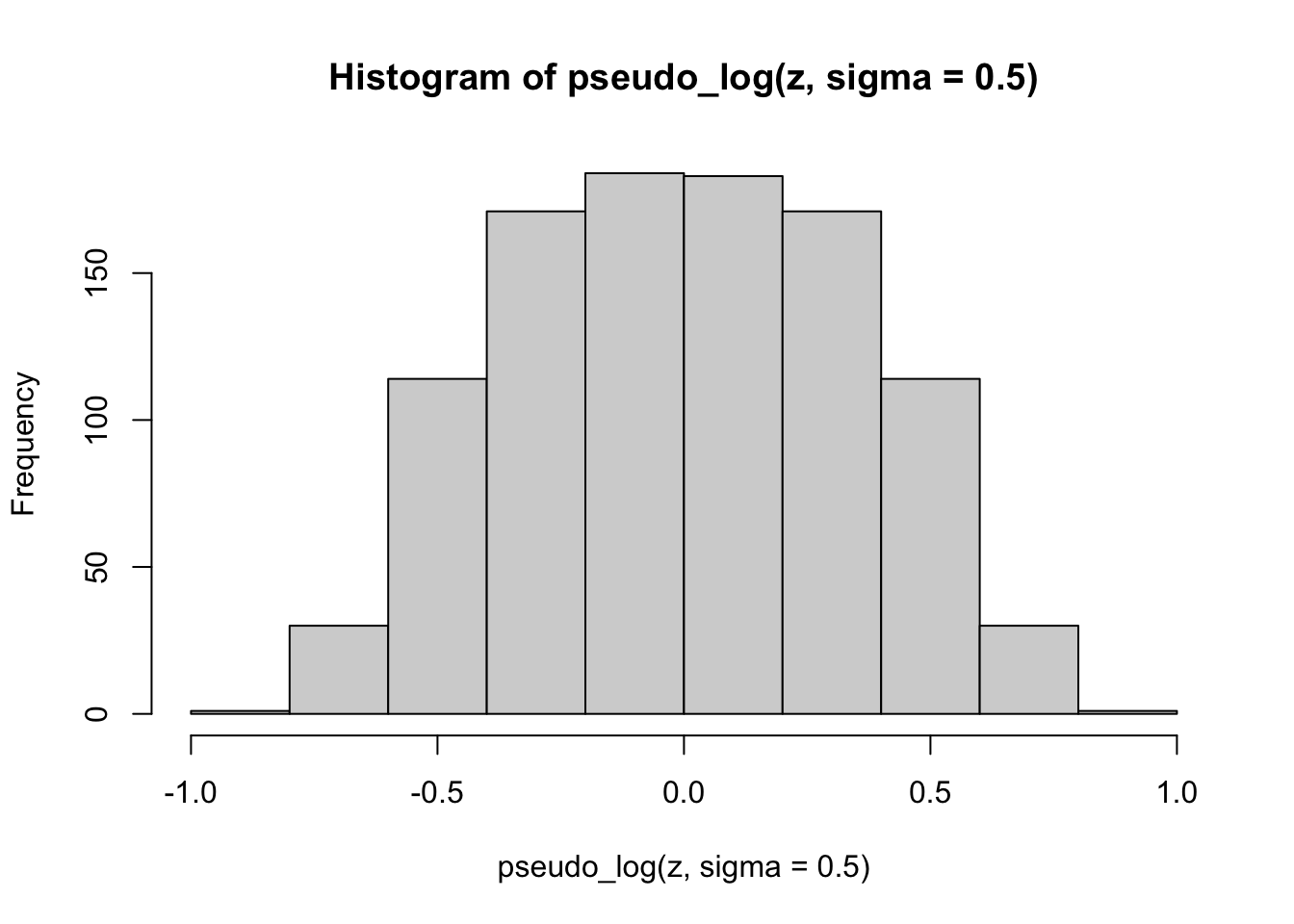
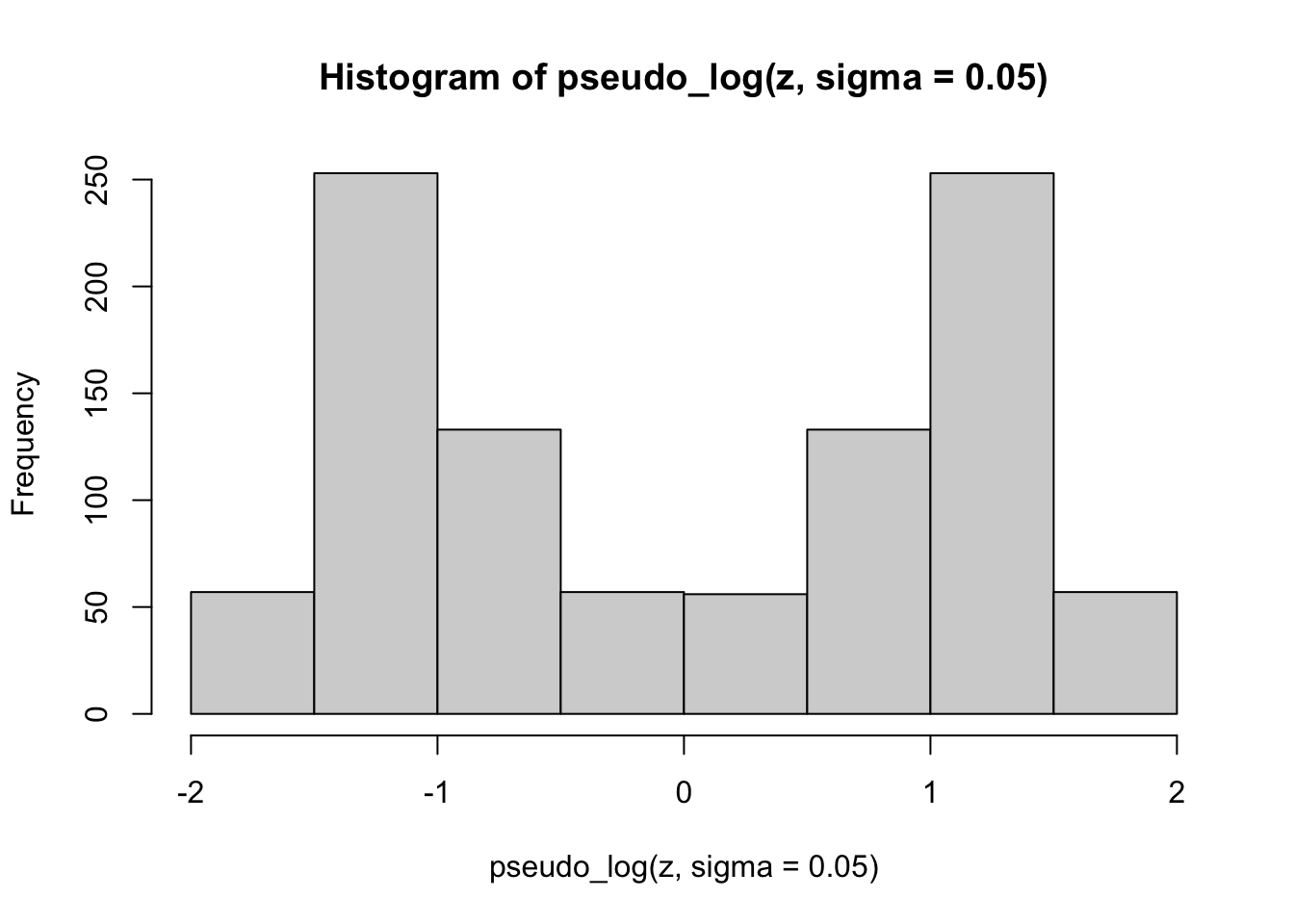
Finally, we apply the pseudo-logarithmic transformation to the original normal deviates. We learn that a higher value for the parameter \(\sigma\) makes the distribution ‘slimmer’ while a lower value makes it ‘fatter’, and it is even possible to induce bimodality with low values of sigma:
z <- qnorm(p, mean = 0, sd = 1)
hist(z)
hist(pseudo_log(z, sigma = 1))
hist(pseudo_log(z, sigma = 2))
hist(pseudo_log(z, sigma = 0.5))
hist(pseudo_log(z, sigma = 0.05))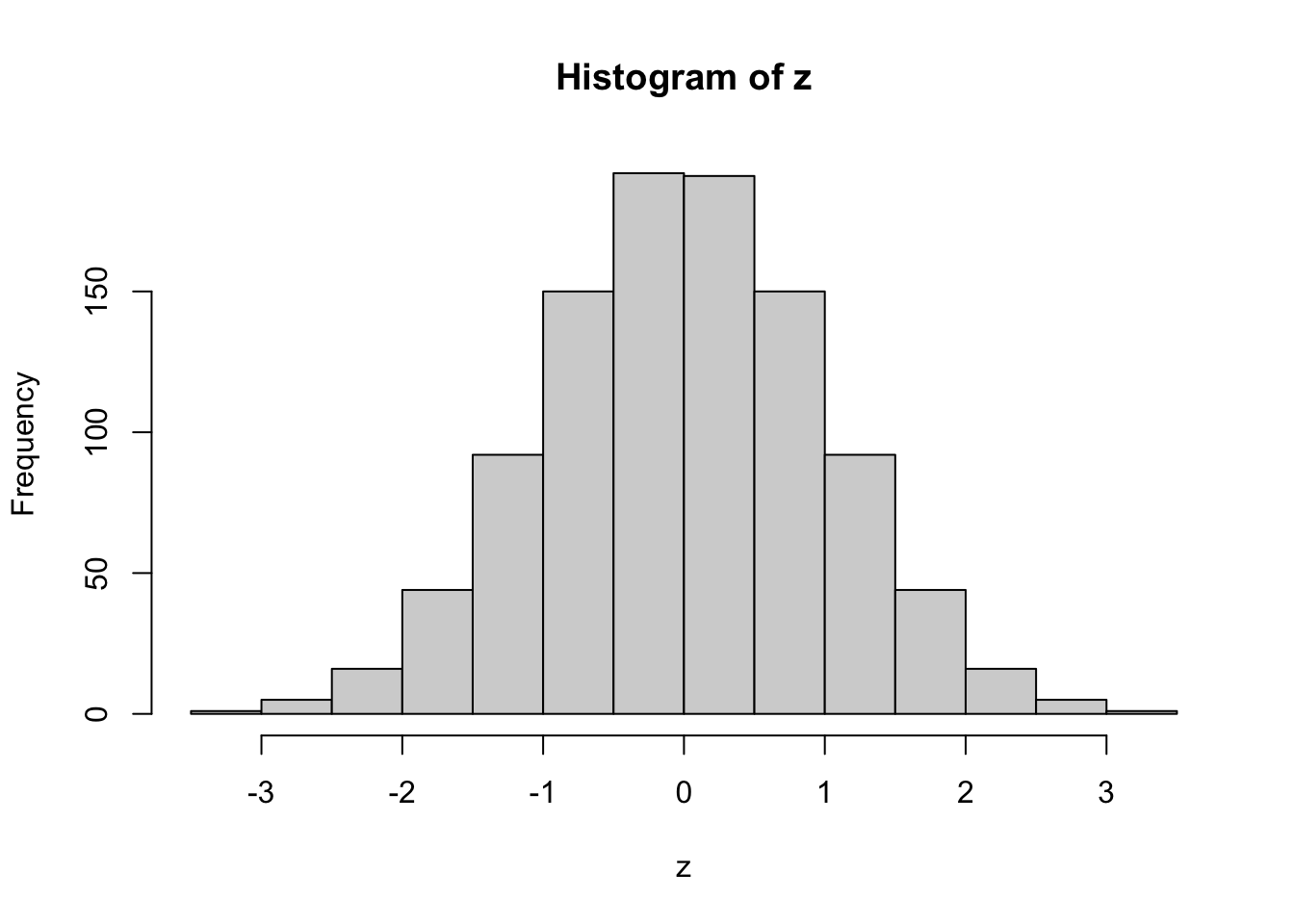
Any test statistic for testing normality could be chosen to find a suitable value of \(\sigma\) that induces normality into the transformed values. Here we use the (Pearson) correlation coefficient to compare the empirical distribution with normal deviates.
We simulate from a shifted log normal distribution and evaluate the value of \(\sigma\) that optimizes agreement with a normal distribution:
# #| layout-ncol: 2 #Remark. did not render nicely in docx (GH)
x <- sort(exp(rnorm(1000) + 3))
hist(x)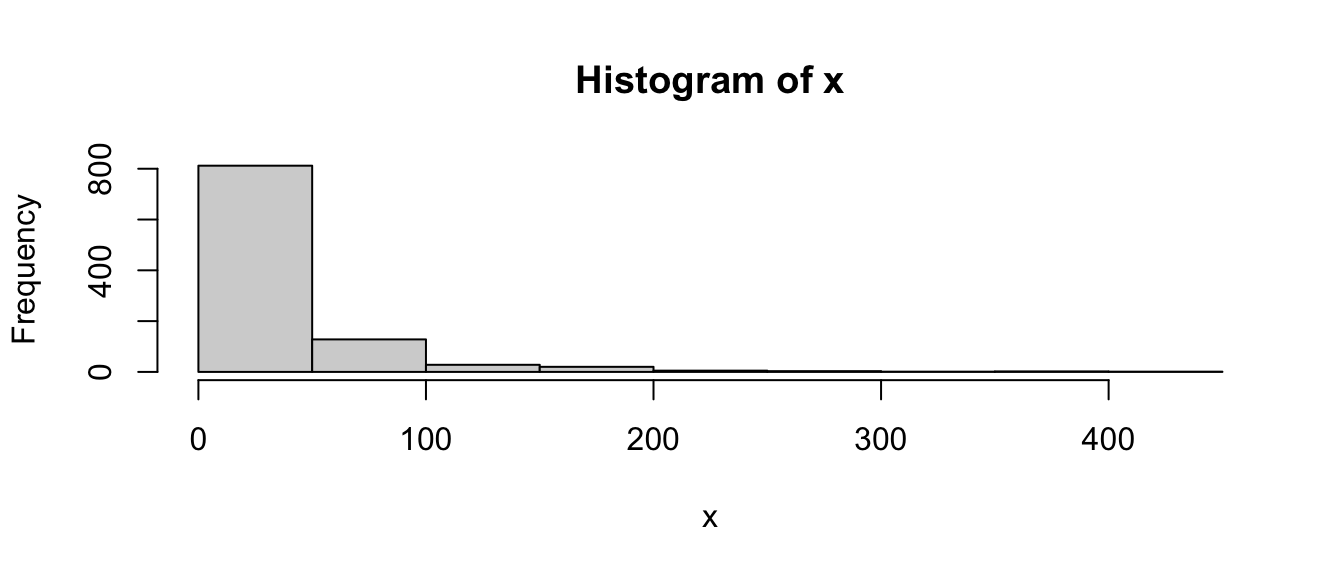
sigmas <- 2 ** seq(-10, 10, 1)
origcor <- cor(qnorm((1:length(x) - 0.5) / length(x)), x)
ncorx <-
sapply(sigmas, function(X)
cor(qnorm((1:length(
x
) - 0.5) / length(x)), pseudo_log(x, X)))
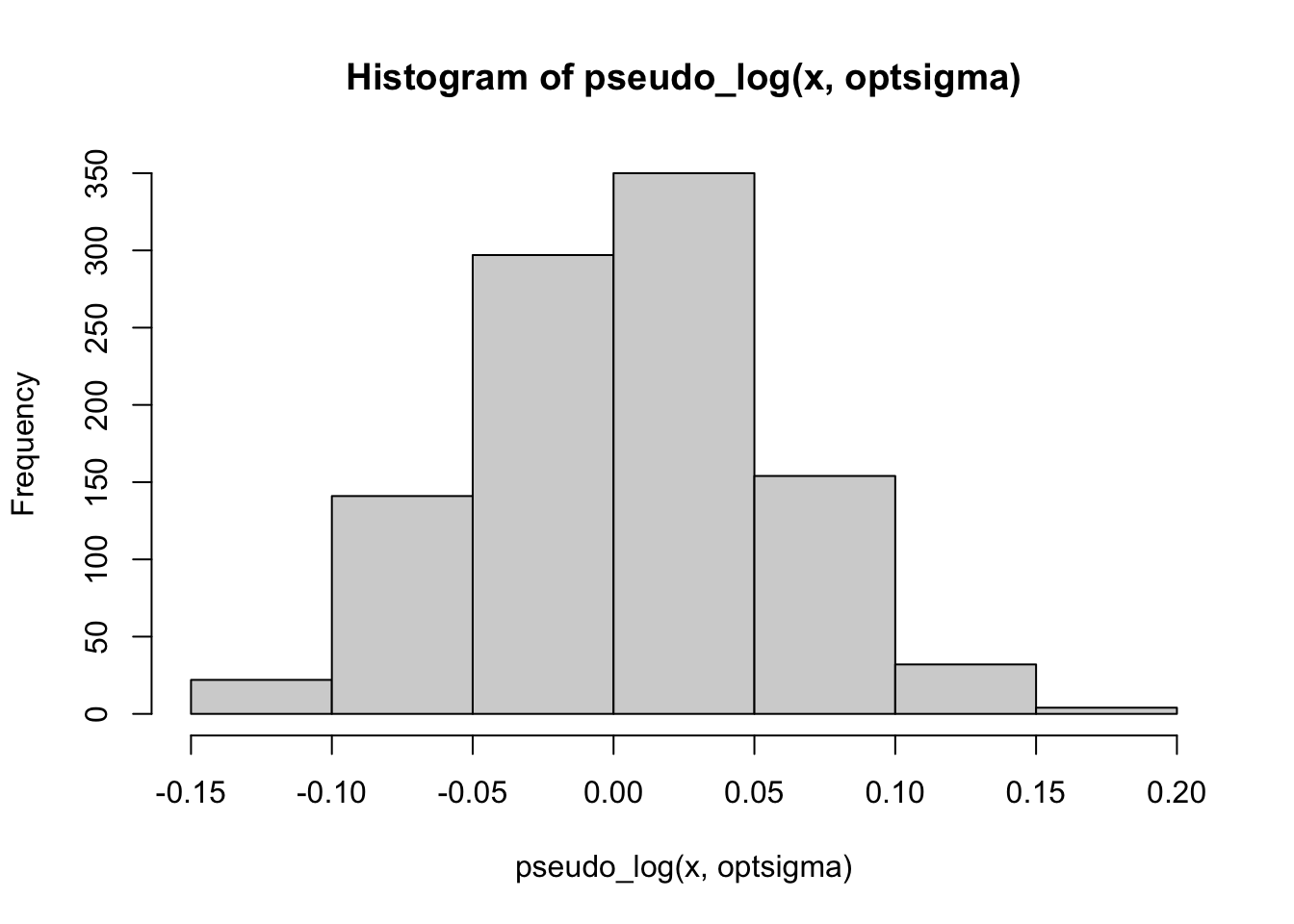
cat("Optimal sigma: ")Optimal sigma: (optsigma <- sigmas[ncorx == max(ncorx)])[1] 0.25plot(log2(sigmas), ncorx, ylab = "Correlation with normal deviates", ylim =
c(0, 1))
points(log2(sigmas)[ncorx == max(ncorx)], max(ncorx), pch = 19)
abline(h = origcor)
legend(
"bottomright",
lty = c(1, NA, NA),
pch = c(NA, 1, 19),
legend = c("Original", "Pseudo-log", "Pseudo-log with optimal sigma")
)
box()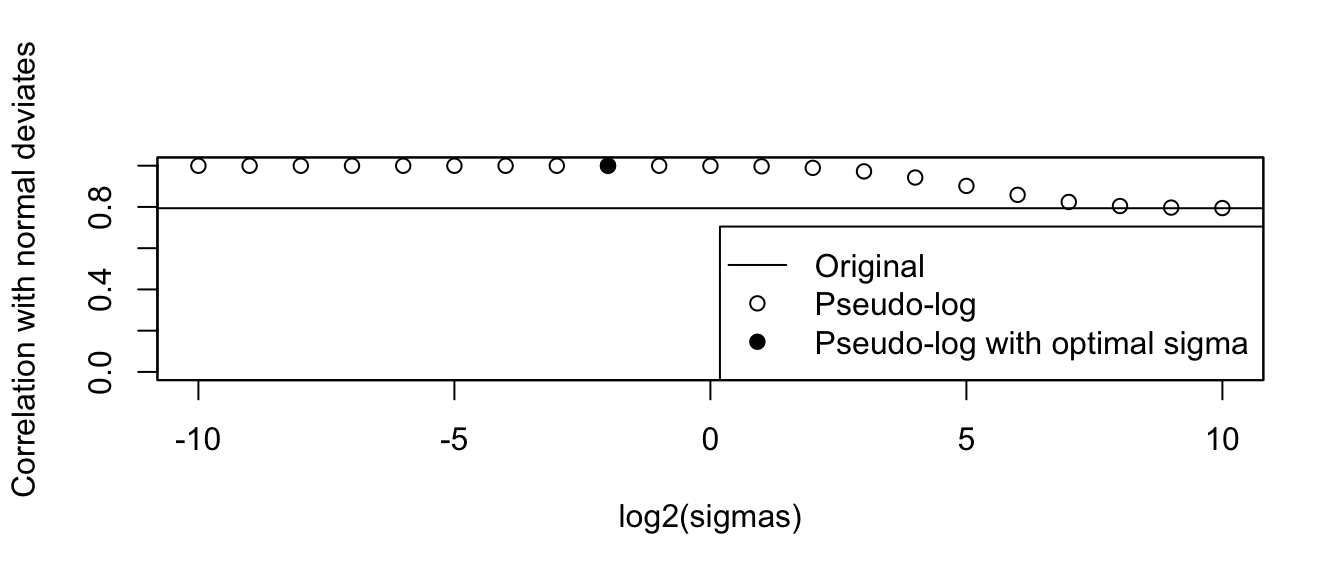
hist(pseudo_log(x, optsigma))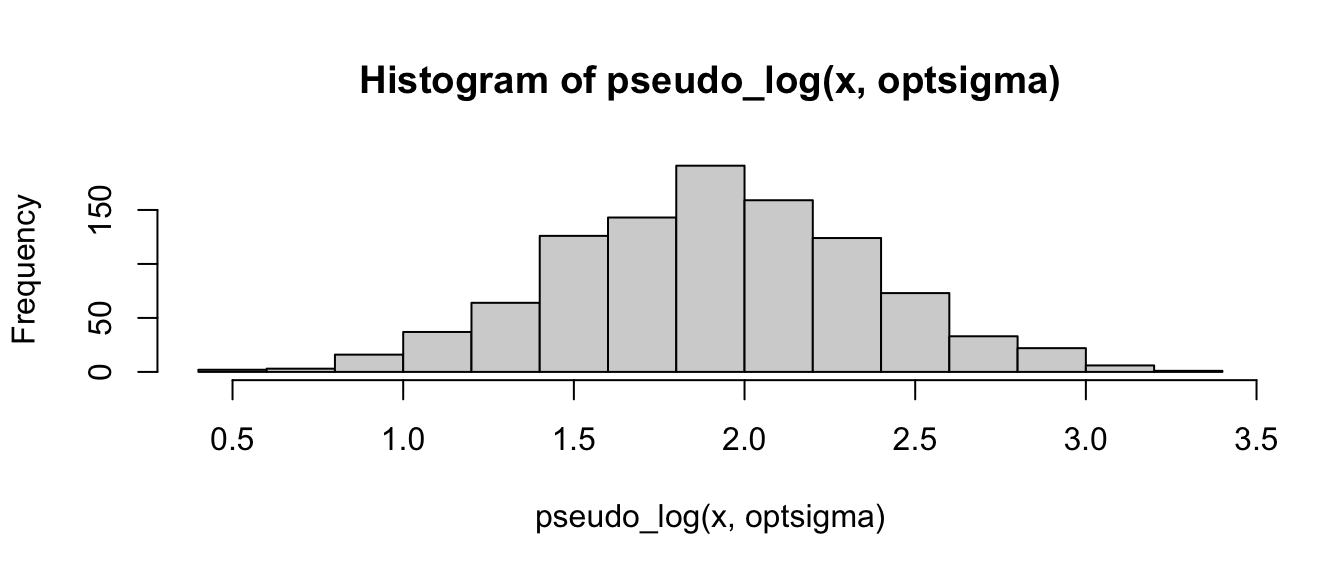
Also with an exponential distribution, the pseudo-logarithm may achieve a better agreement with a normal:
x <- sort(exp(rnorm(1000) + 3))
hist(x)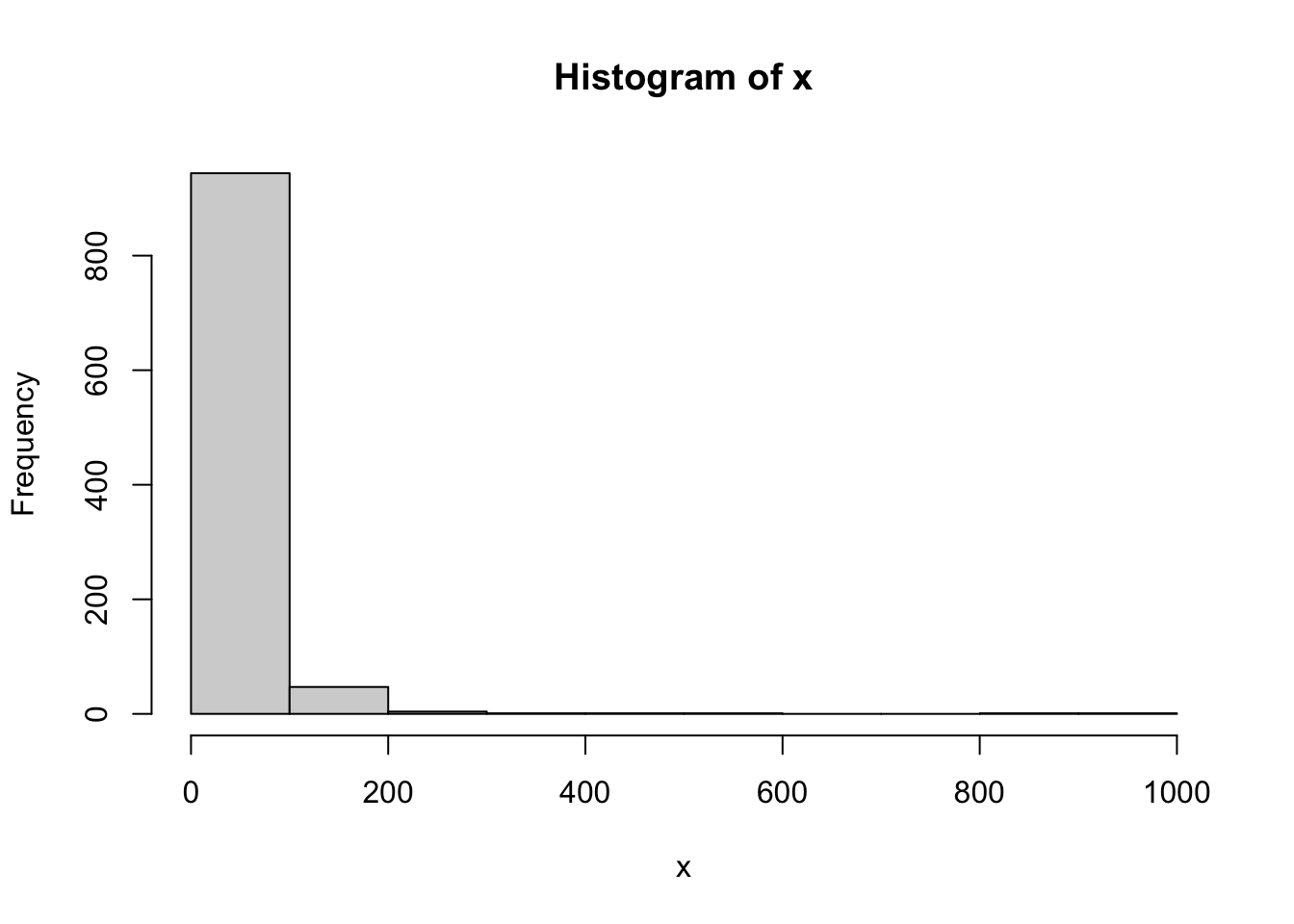
sigmas <- 2 ** seq(-10, 10, 1)
origcor <- cor(qnorm((1:length(x) - 0.5) / length(x)), x)
ncorx <-
sapply(sigmas, function(X)
cor(qnorm((1:length(
x
) - 0.5) / length(x)), pseudo_log(x, X)))
cat("Optimal sigma: ")Optimal sigma: (optsigma <- sigmas[ncorx == max(ncorx)])[1] 0.0009765625plot(log2(sigmas), ncorx, ylab = "Correlation with normal deviates", ylim =
c(0, 1))
points(log2(sigmas)[ncorx == max(ncorx)], max(ncorx), pch = 19)
abline(h = origcor)
legend(
"bottomright",
lty = c(1, NA, NA),
pch = c(NA, 1, 19),
legend = c("Original", "Pseudo-log", "Pseudo-log with optimal sigma")
)
box()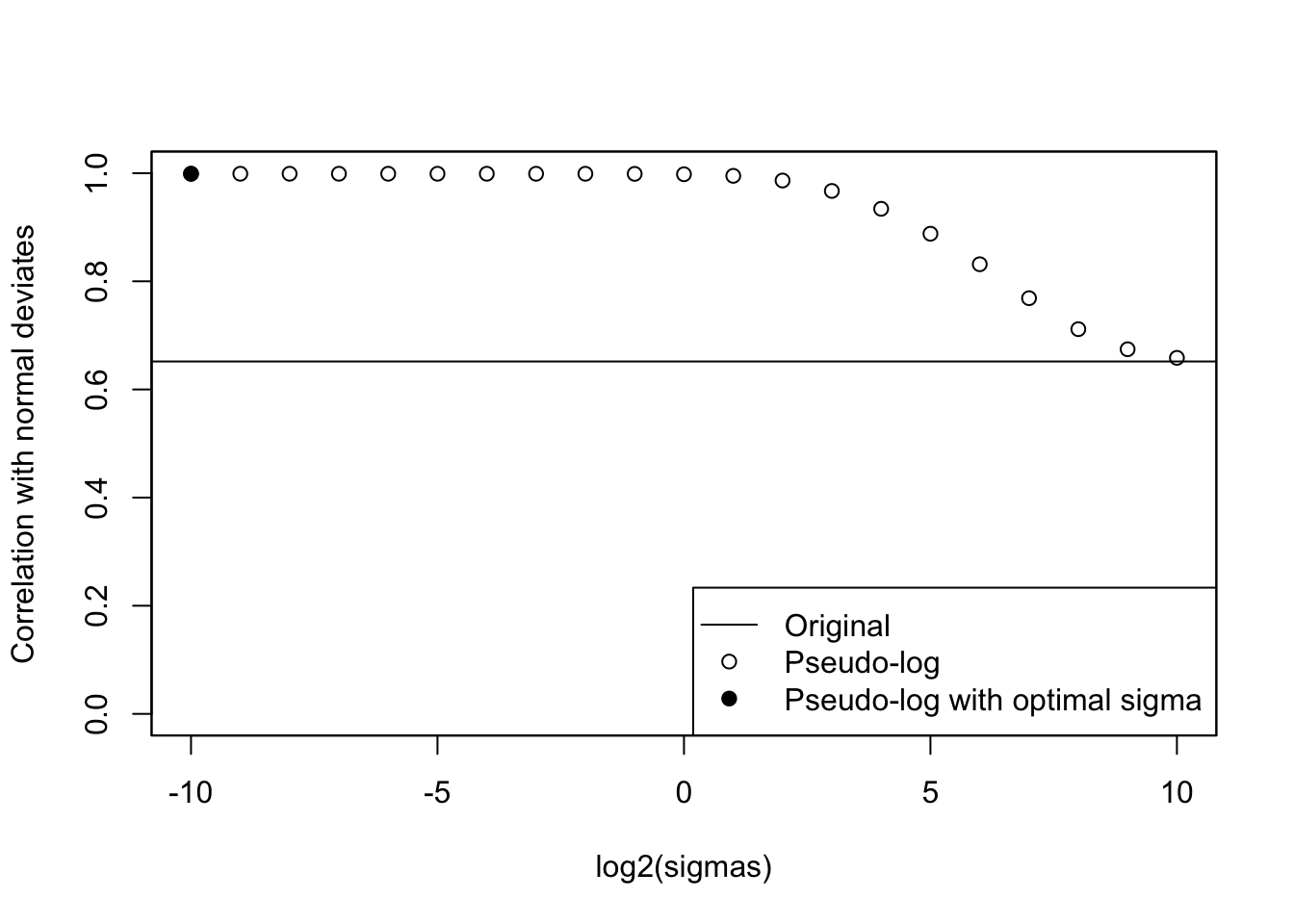
hist(pseudo_log(x, optsigma))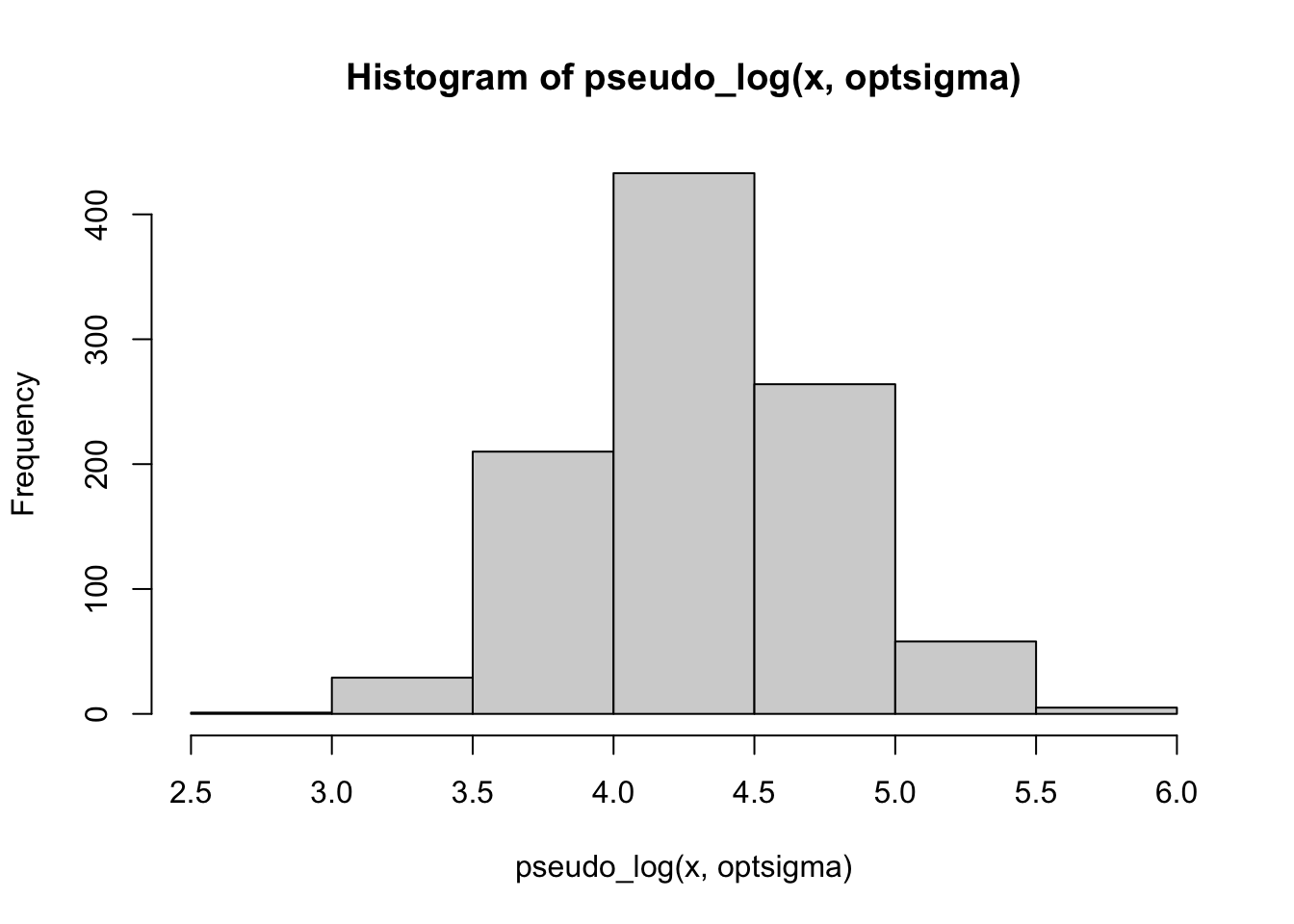
Now we mix a normal, lognormal and exponential distribution:
x1 <- scale(rnorm(1000))
x2 <- scale(rexp(1000))
x3 <- scale(exp(rnorm(1000)))
p1 <- rbinom(1000, 1, 0.33)
p2 <- rbinom(1000, 1, 0.5)
x <- p1 * x1 + (1 - p1) * (p2 * x2 + (1 - p2) * x3)
x <- x - min(x)
x <- sort(x)
hist(x)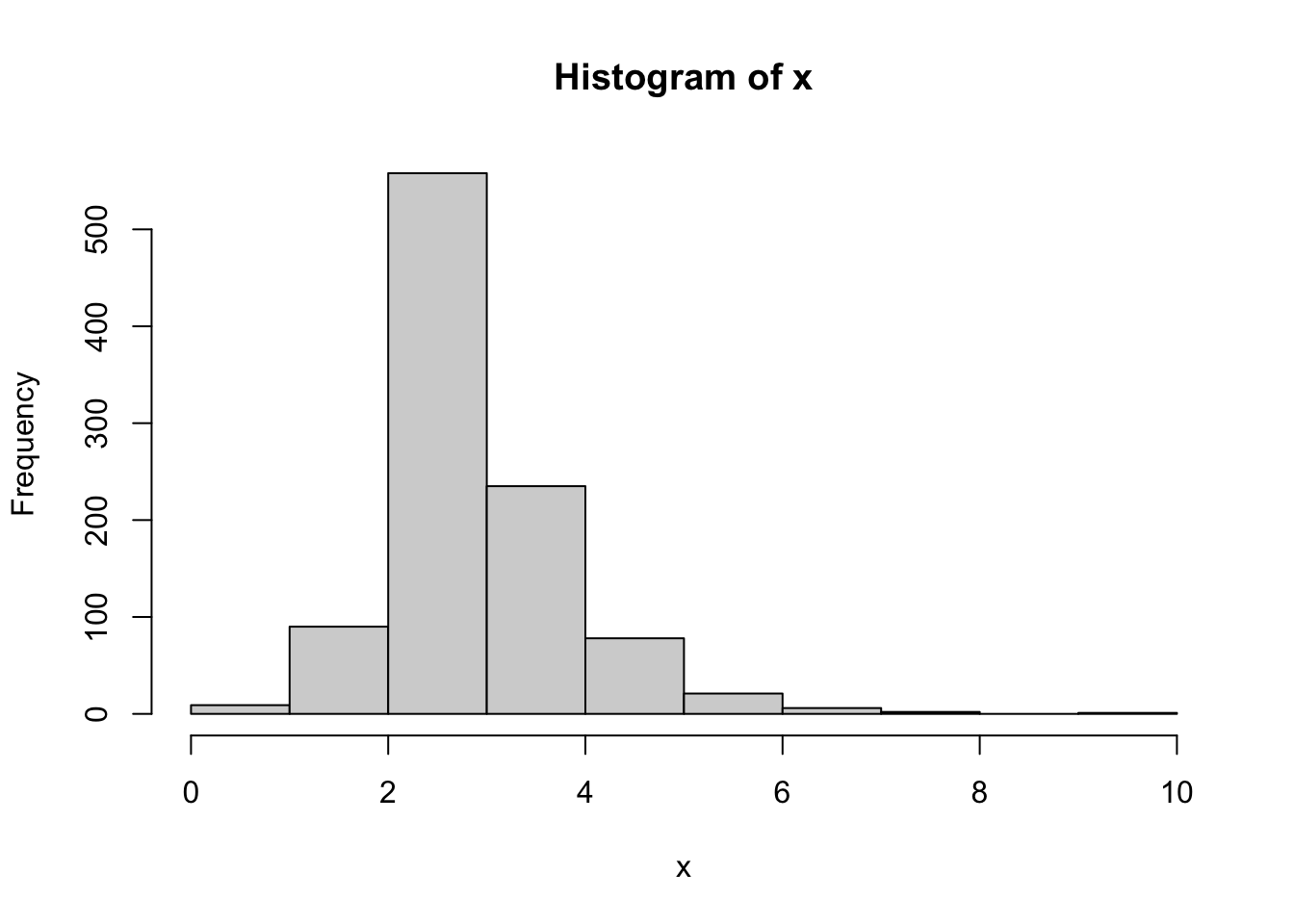
sigmas <- 2 ** seq(-10, 10, 1)
origcor <- cor(qnorm((1:length(x) - 0.5) / length(x)), x)
ncorx <-
sapply(sigmas, function(X)
cor(qnorm((1:length(
x
) - 0.5) / length(x)), pseudo_log(x, X)))
cat("Optimal sigma: ")Optimal sigma: (optsigma <- sigmas[ncorx == max(ncorx)])[1] 1plot(log2(sigmas), ncorx, ylab = "Correlation with normal deviates", ylim =
c(0, 1))
points(log2(sigmas)[ncorx == max(ncorx)], max(ncorx), pch = 19)
abline(h = origcor)
legend(
"bottomright",
lty = c(1, NA, NA),
pch = c(NA, 1, 19),
legend = c("Original", "Pseudo-log", "Pseudo-log with optimal sigma")
)
box()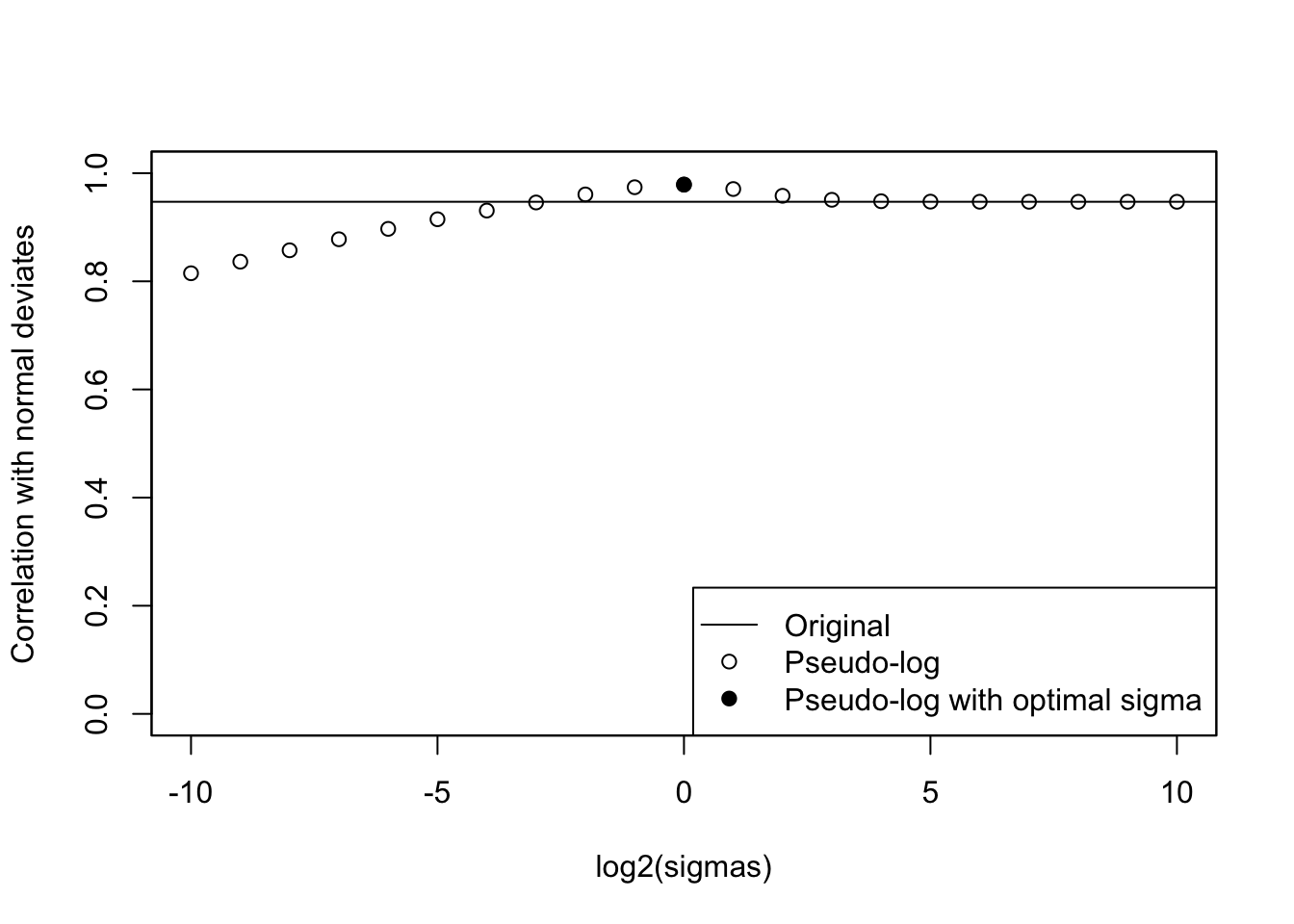
hist(pseudo_log(x, optsigma))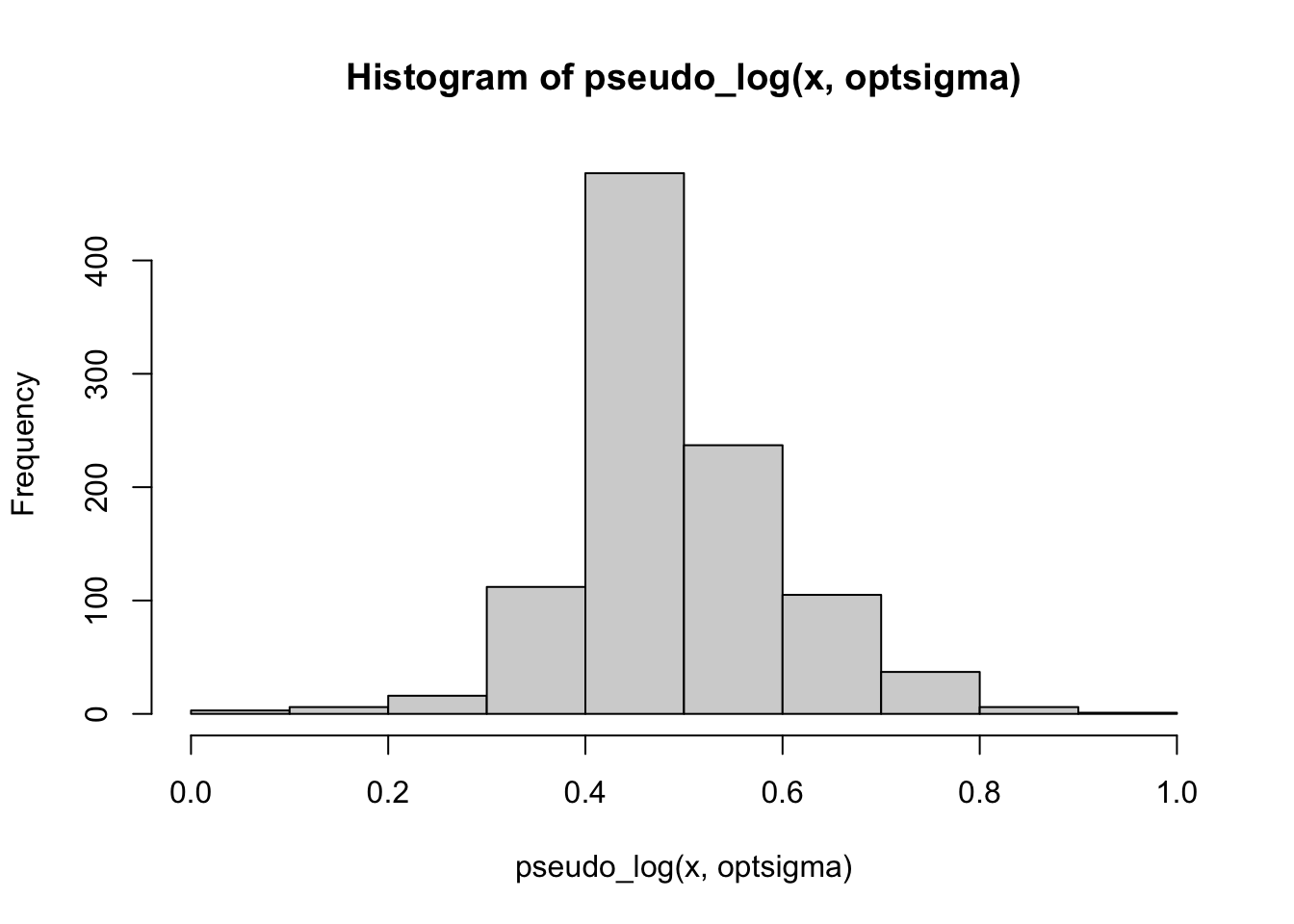
With simulated normal deviates, the pseudo-logarithm cannot improve the already perfect normality.
x <- sort(rnorm(1000))
hist(x)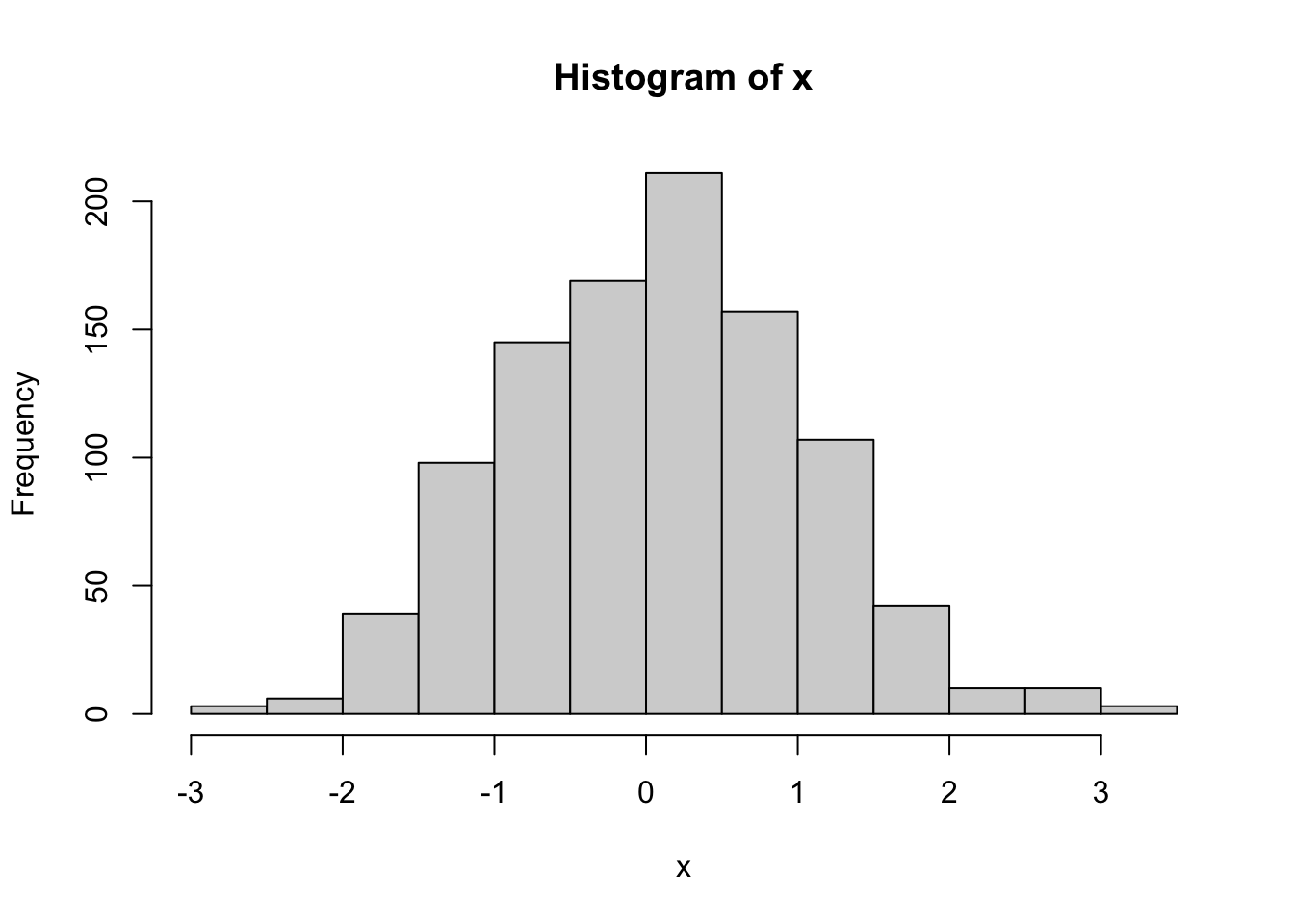
sigmas <- 2 ** seq(-10, 10, 1)
origcor <- cor(qnorm((1:length(x) - 0.5) / length(x)), x)
ncorx <-
sapply(sigmas, function(X)
cor(qnorm((1:length(
x
) - 0.5) / length(x)), pseudo_log(x, X)))
cat("Optimal sigma: ")Optimal sigma: (optsigma <- sigmas[ncorx == max(ncorx)])[1] 4plot(log2(sigmas), ncorx, ylab = "Correlation with normal deviates", ylim =
c(0, 1))
points(log2(sigmas)[ncorx == max(ncorx)], max(ncorx), pch = 19)
legend(
"bottomright",
lty = c(1, NA, NA),
pch = c(NA, 1, 19),
legend = c("Original", "Pseudo-log", "Pseudo-log with optimal sigma")
)
abline(h = origcor)
box()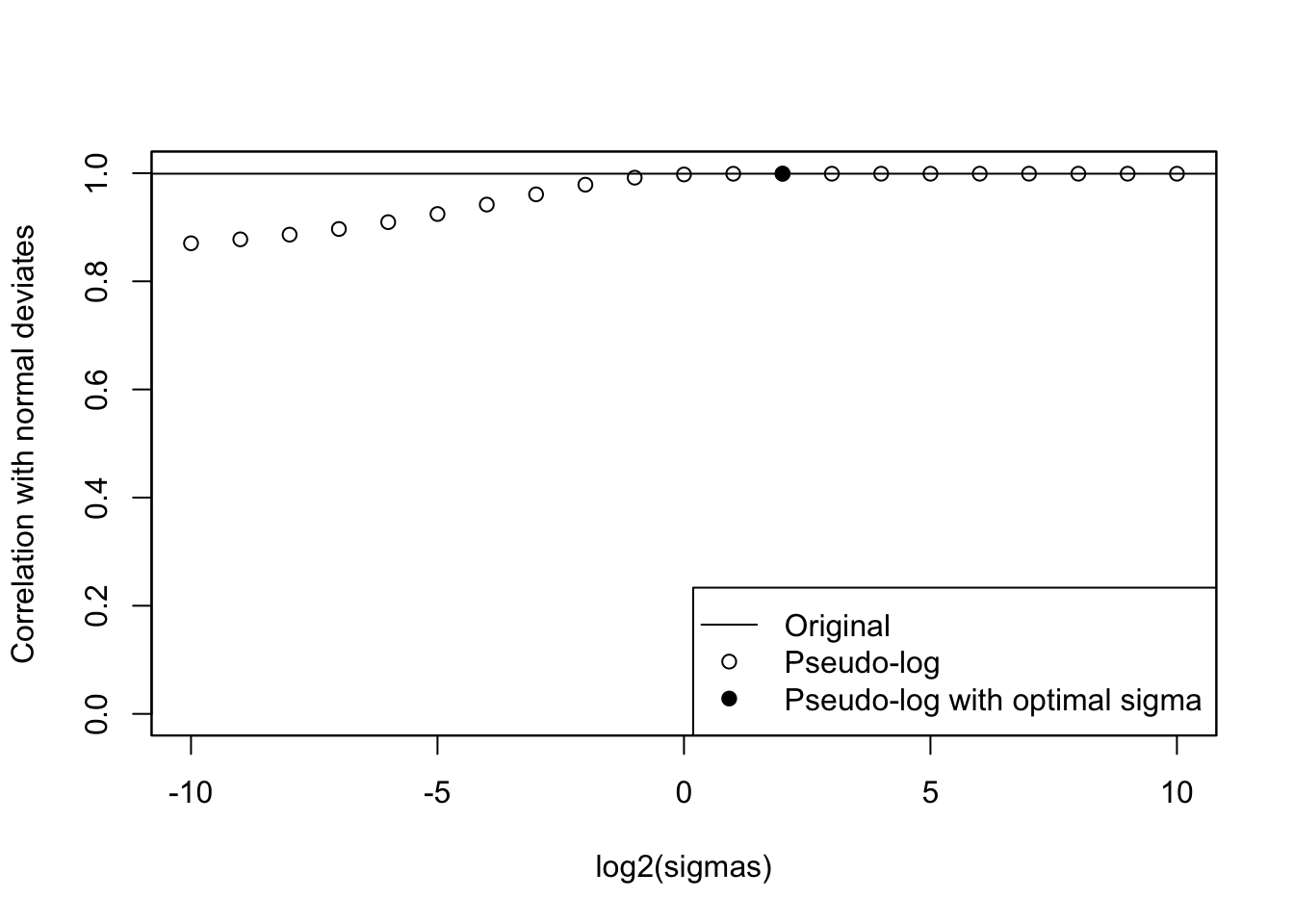
hist(pseudo_log(x, optsigma))